 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr036250.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 427aa MW: 48870.7 Da PI: 6.3082 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 86.5 | 3.2e-27 | 73 | 154 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W ++e++ Li++rr m+ ++++k++k+lWe++s+kmre+gf rsp++C++kw+nl k++kk k+++++ s +++y++++e
Pbr036250.1 73 WVQDETRSLIALRRGMDGLFNTSKSNKHLWEQISAKMREKGFDRSPTMCTDKWRNLLKEFKKAKHHDRGS---GSAKMSYYKEIE 154
********************************************************************84...556899***997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 8.211 | 65 | 129 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-4 | 65 | 134 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 0.0012 | 69 | 131 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.24E-29 | 71 | 136 | No hit | No description |
| Pfam | PF13837 | 7.4E-21 | 72 | 156 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MYLPEKARPI DFYKEEASRD MMMEVVVSNE DLPQSQSQSQ AHLQQPLPQQ MLMGDSSGED 60 HEVRAPKKRA ETWVQDETRS LIALRRGMDG LFNTSKSNKH LWEQISAKMR EKGFDRSPTM 120 CTDKWRNLLK EFKKAKHHDR GSGSAKMSYY KEIEEILKDR SKNTQYKSPT PPKVDSFVQF 180 SDKDNGGDLS FDLPCFNVPY QERNRPCNLR RTAAQIRNFF DGFSAHAGIE DASISFAPVE 240 ASGRPTLNLE RRLDHDGHPL AITAADAVTA GGVPPWNWRE TPGNGVEGQA YGGRVILVNW 300 GDYTRRIGID GTADAIKDAI KSAFRLRTKR SFWLEDEDQI VRSLDRDMPL GNYTLHLDEG 360 VAIKVCLYDE SDHIPVHTEE KLFFTEEDYR EFLSRRGWSY LREFDGYRNI ENMDDLRPGA 420 IYRGVS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 3e-53 | 67 | 152 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00004 | PBM | Transfer from 920224 | Download |
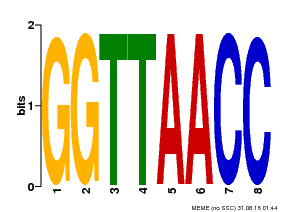
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr036250.1 |
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009359290.1 | 0.0 | PREDICTED: trihelix transcription factor GT-1 isoform X2 | ||||
| Swissprot | Q9FX53 | 0.0 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A498HL35 | 0.0 | A0A498HL35_MALDO; Uncharacterized protein | ||||
| STRING | XP_009359289.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6817 | 32 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 0.0 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



