 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pbr036965.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Maleae; Pyrus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1464aa MW: 163496 Da PI: 8.7802 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 10.7 | 0.0017 | 1347 | 1372 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C C++sF +k++L H r+ +
Pbr036965.1 1347 FECDfeGCTMSFASKHDLSLHKRNiC 1372
789999****************9877 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00034 | 1372 | 1394 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Pbr036965.1 1372 CPvkGCGKKFFSHKYLVQHRRVH 1394
9999*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 2.9E-15 | 24 | 65 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.605 | 25 | 66 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 3.7E-15 | 26 | 59 | IPR003349 | JmjN domain |
| SMART | SM00558 | 1.9E-49 | 200 | 369 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 32.697 | 203 | 369 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 2.09E-25 | 214 | 384 | No hit | No description |
| Pfam | PF02373 | 2.0E-36 | 233 | 352 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 7.0E-4 | 1347 | 1369 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 16 | 1347 | 1369 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0045 | 1370 | 1394 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.152 | 1370 | 1399 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1372 | 1394 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.8E-7 | 1372 | 1398 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.27E-9 | 1386 | 1428 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-7 | 1399 | 1424 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.637 | 1400 | 1429 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0017 | 1400 | 1424 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1402 | 1424 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.7E-7 | 1418 | 1452 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-8 | 1425 | 1453 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.221 | 1430 | 1461 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.7 | 1430 | 1456 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1432 | 1456 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1464 aa Download sequence Send to blast |
MAASSGLIAE ANPEVFPWLK TLPVAPEYHP TWAEFQDPIA YIFKIEKEAS QYGICKIVPP 60 VPPSTKKTTI GNLNRSLAAR AGVSGSSCPK SEPTFTTRQQ QIGFCPRRPR PVHRPVWQSG 120 EQYTFQEFEV KAKSFEKSYL RKCSKKGGLS PLEIETLYWK ATVDKPFSVE YANDMPGSAF 180 MPVGARKSSS TSRDAGDNVT LGETAWNMRG VSRSKGSLLR FMKEEIPGVT SPMVYIAMLF 240 SWFAWHVEDH DLHSLNYLHM GAGKTWYGVP KDAAVAFEEV VRVQGYAGEI NPLVTFSTLG 300 EKTTVMSPEV FVSSGIPCCR LVQNAGEFVV TFPRAYHTGF SHGFNCAEAA NIATPEWLRV 360 AKDAAIRRAS INYPPMVSHF QLLYDLALAL CSRMPSRISS EPRSSRLKDK RKCEGDIVVK 420 ELFVQDVIQN NDLLNVLGKG SPIVLLPQSS SDLSVCSKLR AGSQSRVNPG FSQGSHSLRE 480 EMKSSGSVSD GLMIDRQQVK GFYSVRGKLA SPSESNTLPS LSGSNNVRGL NSKRSNLNCE 540 RESNVKGEGL SDQRLFSCVT CGILSFACVA IIQPTEEAAR YLMSADCSFF NDWVVGSGLA 600 SEVFPVATGD PITSKNAPCT GLEEYNAPAG LYDVLVQSGD CQIQVVDQGN EMVSNTEMPR 660 ETSALGLVAL NYGNSSDSEE DQVEPDVPVC SDEPNMTNCS LESRYRDQSA SPPWRNPYAG 720 TSGVHSPSSQ GSGCENELRL QTFDHYATDG RKIANFKDSS LQNFDCSADF KTNNSASTAT 780 GFGKAIVPIQ KKSMSFHPGC DEDSSRMHVF CLEHAVEVEQ QLRSIGGVHI LLLCHPDYPR 840 IEDEAKSMAE ELGISYLWND MAFMNSAKED ETRIQLALDS EEAIAGNGDW AVKLGINLFY 900 SASLSRSHLY SKQMPYNSVI YKAFGRSSPA SSPTRIDAYG RRGGKPKKVV AGKWCGKVWM 960 SNQVHSFLVK RDPEEEVEVA EDEEERTFRA WAMPDEDHEV KSEITRKTEK TVKKYARKRK 1020 MTADTRTTKK ARCFDKEDAV SDYSVDDNST QQQRRSKQAK HTESGRTKKP KHVVTEDAVS 1080 DDSMQDDDSL QQSGRFLHSE QVKYIERSDV SDDSMGVESH QQHKRTAKSK QFKPVETDVV 1140 SDDSFEGSSH QPQRVLRSKT TKCTGRENLI SEDVHGFGSH QQRRSISRSK QARARFIERE 1200 DTALDETRED NFQQHKRILR NKQTKPETRG KMRQETPRQV KQGTAPLVKQ GTRTPRNQQT 1260 PQQTKKQTPR LRNNQSEQNS FDLYAEEETE GGPSTRLRKR APKPSKVPGT KPKEQQQTAR 1320 KKAKNASAGK AQGGRNETKL KEEDAEFECD FEGCTMSFAS KHDLSLHKRN ICPVKGCGKK 1380 FFSHKYLVQH RRVHTDDRPL RCPWKGCKMT FKWAWARTEH IRVHTGARPY VCAEPGCAQT 1440 FRFVSDFSRH KRKTGHSVKK SKK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 6e-74 | 18 | 390 | 8 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 6e-74 | 18 | 390 | 8 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
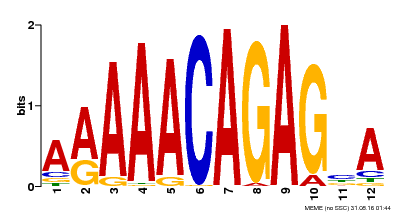
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Pbr036965.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009359508.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6-like | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A498I3J3 | 0.0 | A0A498I3J3_MALDO; Uncharacterized protein | ||||
| STRING | XP_009359508.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5648 | 32 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



