 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00021g00922.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 803aa MW: 87845.8 Da PI: 6.3709 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.6 | 6.7e-21 | 98 | 153 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Peaxi162Scf00021g00922.1 98 KKRYHRHTPQQIQELEALFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 153
678899***********************************************999 PP
| |||||||
| 2 | START | 209.4 | 1.3e-65 | 307 | 532 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT CS
START 1 elaeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdet 75
ela++a++elvk+a+ +ep+W++s e++n +e++++f++ + + +ea+r++g+v+ +++ lve+l+d++ +W e+
Peaxi162Scf00021g00922.1 307 ELALAAMDELVKLAQTDEPLWFRSMegggkEILNHEEYMRTFTPCIGmrpnsFVSEASRETGMVIINSMALVETLMDSN-KWAEM 390
5899***************************************9999****9***************************.***** PP
-S....EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESS CS
START 76 la....kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellp 149
++ + +t++vissg galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvSvd+ ++ + + + +++lp
Peaxi162Scf00021g00922.1 391 FPclipRTSTTDVISSGmggtrnGALQLMHAELQVLSPLVPiREVNFLRFCKQHAEGVWAVVDVSVDNIREASGAPTFPNCRRLP 475
************************************************************************99*********** PP
EEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 150 Sgiliepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
Sg+++++++ng+skvtwveh++++++++h l+rsl++ g+ +ga++wvatlqrqce+
Peaxi162Scf00021g00922.1 476 SGCVVQDMPNGYSKVTWVEHAEYDESVIHHLYRSLISAGMGFGAQRWVATLQRQCEC 532
*******************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-22 | 84 | 155 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.84E-21 | 86 | 155 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.54 | 95 | 155 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.1E-17 | 96 | 159 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.27E-18 | 97 | 155 | No hit | No description |
| Pfam | PF00046 | 2.0E-18 | 98 | 153 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 130 | 153 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.034 | 298 | 535 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.2E-33 | 300 | 532 | No hit | No description |
| CDD | cd08875 | 1.66E-123 | 302 | 531 | No hit | No description |
| SMART | SM00234 | 1.9E-50 | 307 | 532 | IPR002913 | START domain |
| Pfam | PF01852 | 2.8E-57 | 307 | 532 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.68E-24 | 551 | 789 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 803 aa Download sequence Send to blast |
MNFGDFLDNN SGGGGGARIV ADIPFNNNNN SNSSNNSINM PTGAISQPRL LSQSLNKSIN 60 GRRSREEEPE SRSGSDNLEE GASGDDQEDA ADKPPRRKKR YHRHTPQQIQ ELEALFKECP 120 HPDEKQRLEL SKRLCLETRQ VKFWFQNRRT QMKTQLERHE NSLLRQENDK LRAENMSIRE 180 AMRNPICTNC GGPAMIGEIS LEEQHLRIEN ARLKDELDRV CGLAGKFLGR PISSLVTSMP 240 PPMPNSSLEL GVGNNGFVVG GLSNVPTTLP LAPSPDFGVG ISNSLPMVTR QTSTGIERSL 300 ERSMYLELAL AAMDELVKLA QTDEPLWFRS MEGGGKEILN HEEYMRTFTP CIGMRPNSFV 360 SEASRETGMV IINSMALVET LMDSNKWAEM FPCLIPRTST TDVISSGMGG TRNGALQLMH 420 AELQVLSPLV PIREVNFLRF CKQHAEGVWA VVDVSVDNIR EASGAPTFPN CRRLPSGCVV 480 QDMPNGYSKV TWVEHAEYDE SVIHHLYRSL ISAGMGFGAQ RWVATLQRQC ECLAILMSST 540 VSSRDHTAIT PSGRRSMLKL AQRMTNNFCA GVCASTVHKW NKLCAGNVDE DVRVMTRKSV 600 DDPGEPPGIV LSAATSVWLP VSPQRLFDFL RDERLRSEWD ILSNGGPMQE MAHIAKGQDH 660 GNCVSLLRAS AMNTNQSSML ILQETCIDAA GALVVYAPVD IPAMHVVMNG GDSAYVALLP 720 SGFSIVPDGP GSRGPNNANC GSNGPSCNGG PGERISGSLL TVAFQILVNS LPTAKLTVES 780 VETVNNLISC TVQKIKAALH CES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
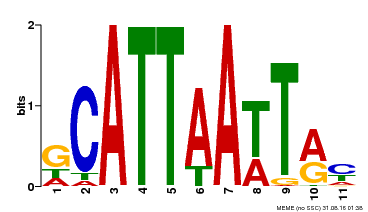
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GQ222185 | 0.0 | GQ222185.1 Solanum lycopersicum cultivar M82 cutin deficient 2 (CD2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019259406.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A1J6ISN2 | 0.0 | A0A1J6ISN2_NICAT; Homeobox-leucine zipper protein anthocyaninless 2 | ||||
| STRING | XP_009782443.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1626 | 24 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



