 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00038g00093.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 809aa MW: 89136.1 Da PI: 6.8908 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59 | 7.7e-19 | 15 | 73 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++ +++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Peaxi162Scf00038g00093.1 15 KYVRYTPEQVEALERLYHDCPKPSSMRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 73
5679*****************************************************97 PP
| |||||||
| 2 | START | 176.1 | 2.1e-55 | 159 | 367 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEEC CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevis 86
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++e+l+v+
Peaxi162Scf00038g00093.1 159 IAEETLTEFLSKATGTAVEWVQMPGMKPGPDSIGIIAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVEVLNVLP 242
789*******************************************************.8888888888**************** PP
TT..EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEE CS
START 87 sg..galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvt 165
++ g+++l +++l+a+++l+p Rdf+ +Ry+ + +g++v++++S++++q+ p+ +++vRae lpSg+li+p+++g+s ++
Peaxi162Scf00038g00093.1 243 TAngGTIELLYMQLYAPTTLAPaRDFWLLRYTTVMDDGSLVVCERSLSNTQNGPSmpqVQNFVRAEILPSGYLIRPCDGGGSIIH 327
9999**************************************************9998899************************ PP
EEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 166 wvehvdlkgrlphwllrslvksglaegaktwvatlqrqce 205
+v+h++l++++++++lr+l++s+++ ++kt+va+l+++++
Peaxi162Scf00038g00093.1 328 IVDHMNLEAWSVPEVLRPLYESSAVLAQKTTVAALRYLRQ 367
***********************************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.516 | 10 | 74 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.2E-16 | 12 | 78 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.52E-17 | 15 | 75 | No hit | No description |
| SuperFamily | SSF46689 | 8.13E-17 | 15 | 78 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 2.1E-16 | 16 | 73 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-18 | 17 | 73 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.29E-6 | 67 | 106 | No hit | No description |
| PROSITE profile | PS50848 | 26.441 | 149 | 377 | IPR002913 | START domain |
| CDD | cd08875 | 1.07E-76 | 153 | 369 | No hit | No description |
| SMART | SM00234 | 2.3E-43 | 158 | 368 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 6.8E-23 | 158 | 363 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 1.32E-37 | 158 | 370 | No hit | No description |
| Pfam | PF01852 | 2.9E-53 | 159 | 367 | IPR002913 | START domain |
| Pfam | PF08670 | 8.4E-51 | 666 | 808 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 809 aa Download sequence Send to blast |
MSSCKDGKGV MDNGKYVRYT PEQVEALERL YHDCPKPSSM RRQQLIRECP ILSNIEPKQI 60 KVWFQNRRCR EKQRKEASRL QAVNRKLTAM NKLLMEENDR LQKQVSHLVC ENGFFRRRSH 120 STPLATKDTS CESVVTSGQH HLTSQHPPRD ASPAGLLSIA EETLTEFLSK ATGTAVEWVQ 180 MPGMKPGPDS IGIIAISHGC TGVAARACGL VGLEPTRVAE ILKDRPSWFR DCRAVEVLNV 240 LPTANGGTIE LLYMQLYAPT TLAPARDFWL LRYTTVMDDG SLVVCERSLS NTQNGPSMPQ 300 VQNFVRAEIL PSGYLIRPCD GGGSIIHIVD HMNLEAWSVP EVLRPLYESS AVLAQKTTVA 360 ALRYLRQIAQ EVSQSNVTNW GRRPAALRAL SQRLSRGFNE ALNGLNDEGW SMLGSDGNDD 420 VTILVNSSPE KMMGLNLSFA NGFSSLSNAV MCAKASMLLQ NVPPAILLRF LREHRSEWAD 480 NNIDAYSAAA IKVGPCSLPG ARVGVGHSPE EAMIPRDMFL LQLCSGMDEN AVGTCAELIF 540 APIDASFADD APLLPSGFRI ISLESGKEAS SPNRTLDLTS ALETGPAENK VANDLQTIGG 600 TSRSVMTIAF QFAFESHMQE NVASMARQYV RSIISSVQRV ALALSPSHLG FHGGLRLPLG 660 TPEAHTLARW ICQSYRCFLG MELLKSNTEG SESILKSLWH HTDAIICCSA KSMPVFTFAN 720 QAGLDMLETT LVALQDITLE KIFDDHGKKN LCTEFPQIMQ QGFACLQGGI CLSSMSRPVS 780 YERAVAWKVM NEEDTPHCIC FMFVNWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
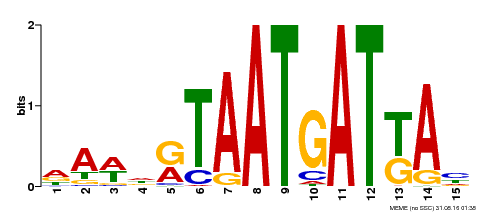
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JQ686925 | 0.0 | JQ686925.1 Nicotiana tabacum cultivar SR1 homeobox-leucine zipper protein ATHB-15-like protein (HB15) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019267274.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15-like isoform X1 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A314KZL5 | 0.0 | A0A314KZL5_NICAT; Homeobox-leucine zipper protein athb-15 | ||||
| STRING | PGSC0003DMT400006735 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA457 | 24 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



