 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00128g00723.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 577aa MW: 63932.7 Da PI: 6.1668 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 77.3 | 1.9e-24 | 191 | 244 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++Wt +LH++Fv+av+q+ G +k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Peaxi162Scf00128g00723.1 191 KARVVWTVDLHQKFVKAVNQI-GFDKVGPKKILDLMGVPWLTRENVASHLQKYRL 244
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 80 | 7.9e-27 | 17 | 125 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgee 83
vl+vdD+p+ +++l+++l+k y ev+ + ++eal+ll+e++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge
Peaxi162Scf00128g00723.1 17 VLVVDDDPTWLKILEKMLKKCSY-EVTICGLAREALHLLRERKdgFDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGET 99
89*********************.***************999999**********************6644.8************ PP
HHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 84 edalealkaGakdflsKpfdpeelvk 109
+ + + ++ Ga d+l Kp+ ++el++
Peaxi162Scf00128g00723.1 100 SRVMKGVQHGACDYLLKPIRMKELRN 125
***********************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 7.68E-36 | 14 | 138 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 1.7E-44 | 14 | 162 | No hit | No description |
| SMART | SM00448 | 5.4E-32 | 15 | 127 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.719 | 16 | 131 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 6.5E-24 | 17 | 126 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 7.80E-31 | 18 | 131 | No hit | No description |
| SuperFamily | SSF46689 | 5.29E-18 | 188 | 248 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-28 | 190 | 249 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.0E-22 | 191 | 244 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.8E-8 | 193 | 243 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 577 aa Download sequence Send to blast |
MMENSKNSGG FPAGLRVLVV DDDPTWLKIL EKMLKKCSYE VTICGLAREA LHLLRERKDG 60 FDIVISDVNM PDMDGFKLLE HVGLEMDLPV IMMSVDGETS RVMKGVQHGA CDYLLKPIRM 120 KELRNIWQHV LRKKMQEVRD IGSLEMDQCD EVWMLNANEV LSGKKRKEFD NKNDEKATSD 180 SRCIDSPCMK KARVVWTVDL HQKFVKAVNQ IGFDKVGPKK ILDLMGVPWL TRENVASHLQ 240 KYRLYLTRLQ KEDEVKASFC GVKHPDVSSK ETSSSLSLQN PVDVCTDVSS DKYGGVSGDK 300 TIVQNGKSNI HESKVNGVVS MPVAEPSSVV GDNFDAQTAG SKINVNNCFG LVTDVKSAKV 360 TTPYCFTGEA PQPQYKQDFK PHFESQNELS HQPLPELPHQ LPVDCTRPTG FVNILPSREE 420 KERPPGTENK ASFYKSRSTR VGKVSPVEST VNLFSFQPVG HPTNFHALGQ IPSTTWSTTS 480 HSVDQILVNG LQSSQGNLTS GSGAVVASAG EDMCGGSIQG ECFPAYTGLR IVEPFGYNDP 540 QPLSGVPTCL YDTLRFDYEY PIDSLEGTVL DQGLFIV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 3e-17 | 190 | 249 | 4 | 63 | ARR10-B |
| Search in ModeBase | ||||||
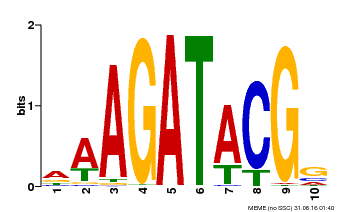
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006341767.1 | 0.0 | PREDICTED: two-component response regulator ARR11 isoform X1 | ||||
| Refseq | XP_006341768.1 | 0.0 | PREDICTED: two-component response regulator ARR11 isoform X2 | ||||
| TrEMBL | A0A0V0IG20 | 0.0 | A0A0V0IG20_SOLCH; Putative two-component response regulator ARR11-like | ||||
| STRING | PGSC0003DMT400031260 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA6788 | 24 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-114 | response regulator 11 | ||||



