 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00129g01043.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 284aa MW: 31965 Da PI: 4.5396 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.2 | 9e-21 | 64 | 117 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t+eq++ Le+ Fe ++++ +++ +LAkklgL+ rqV vWFqNrRa++k
Peaxi162Scf00129g01043.1 64 KKRRLTPEQVHLLEKSFETENKLEPDRKSQLAKKLGLQPRQVAVWFQNRRARWK 117
5678*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 136.4 | 9.3e-44 | 63 | 155 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeve 85
ekkrrl+ eqv+lLE+sFe+e+kLep+rK++la++LglqprqvavWFqnrRAR+ktkqlE+dy++Lk++yd+l ++ ++++k+++
Peaxi162Scf00129g01043.1 63 EKKRRLTPEQVHLLEKSFETENKLEPDRKSQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDHLKSSYDSLLSDFDSITKDND 147
69*********************************************************************************** PP
HD-ZIP_I/II 86 eLreelke 93
+L++e+++
Peaxi162Scf00129g01043.1 148 KLKAEVAS 155
***99875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.52E-20 | 49 | 121 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.236 | 59 | 119 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.9E-20 | 62 | 123 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 6.9E-18 | 64 | 117 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.90E-20 | 64 | 120 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-22 | 65 | 126 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 3.1E-6 | 90 | 99 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 94 | 117 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 3.1E-6 | 99 | 115 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.7E-17 | 119 | 161 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 284 aa Download sequence Send to blast |
MESGRIFFDP SCNGNMLFLG NGDSIFRGAR SMMKMEDSSK RRPFFTSPED LYDEEYYDEQ 60 SPEKKRRLTP EQVHLLEKSF ETENKLEPDR KSQLAKKLGL QPRQVAVWFQ NRRARWKTKQ 120 LERDYDHLKS SYDSLLSDFD SITKDNDKLK AEVASLLEKL QEKVVGSGAG TSEKSEGLEV 180 DAAMTLQVKA EDRLSSGSGG SAVVDEHSPQ LVDSGDSYVH SGHEEYAGVG GYNIPPPVDG 240 LNSEEDDGSD DGSCHGYFSN VFVAAEEHHH HEEEEGEPLG WFWS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 111 | 119 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
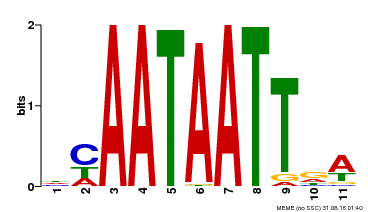
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ808758 | 0.0 | KJ808758.1 Nicotiana tabacum homeodomain leucine zipper protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006338548.1 | 1e-168 | PREDICTED: homeobox-leucine zipper protein HAT5 | ||||
| Swissprot | Q02283 | 4e-94 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | M0ZIA1 | 1e-166 | M0ZIA1_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400001322 | 1e-167 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7891 | 23 | 30 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 4e-78 | homeobox 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



