 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00188g01023.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 285aa MW: 31678.2 Da PI: 4.7198 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 16.9 | 1.2e-05 | 17 | 50 | 18 | 55 |
HHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 18 safeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
s+f+ Lr l+P+ ++K + a+ L ++++Y+ sLq
Peaxi162Scf00188g01023.1 17 SRFQILRNLIPHT----DQKRDTASFLLEVIQYVESLQ 50
68**********8....8*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 3.06E-5 | 17 | 54 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 7.1E-10 | 18 | 68 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-8 | 18 | 71 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MPEHNFYTLD KACLSISRFQ ILRNLIPHTD QKRDTASFLL EVIQYVESLQ EKVQKYEGSY 60 EPWSSEPTKL MPWRNSHWRT QSVPAHPQAL KNDAGTGPAY AGRFDENLAT ATSSMQAIQQ 120 NPVESLPGTD VYFKSIDQQK ELASRGIAIQ MPLQASMPVS LQNDSAFSDS LHRPGSDECP 180 NTTNALNHEE EFMIEGGTIS ISSSYSQGLL DSLTRALQTA GLDLSQATIS LQINLGKQVN 240 SGMTSGASVA KDNETPPRVG NQFMVQFRDE DHDEDLDQAE KRLKI |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
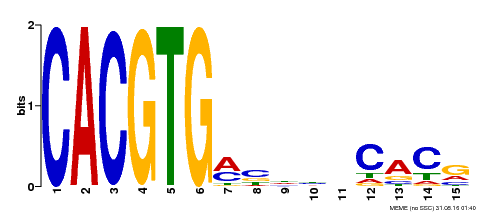
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by heat treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019247319.1 | 1e-134 | PREDICTED: transcription factor BIM2 | ||||
| Swissprot | Q9CAA4 | 1e-74 | BIM2_ARATH; Transcription factor BIM2 | ||||
| TrEMBL | A0A2G3B725 | 1e-134 | A0A2G3B725_CAPCH; Transcription factor BIM2 | ||||
| STRING | XP_009594832.1 | 1e-132 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4426 | 23 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 5e-77 | BES1-interacting Myc-like protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



