 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00241g00423.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 358aa MW: 41391.1 Da PI: 7.1304 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 178.9 | 1.3e-55 | 7 | 136 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgy 82
+ppGfrFhPtdeelv++yL+kkv++++++l +vik+vd+yk+ePwdL++ ++ + e++ewyfFs++dkky+tg+r+nrat++g+
Peaxi162Scf00241g00423.1 7 VPPGFRFHPTDEELVDYYLRKKVTSRRIDL-DVIKDVDLYKIEPWDLQElcRIGTeEQNEWYFFSHKDKKYPTGTRTNRATAAGF 90
69****************************.9***************954555442556************************** PP
NAM 83 WkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrle 129
Wkatg+dk ++s k+ l+g++ktLvfykgrap+g+k+dW+mheyrle
Peaxi162Scf00241g00423.1 91 WKATGRDKGIYS-KHDLIGMRKTLVFYKGRAPNGQKSDWIMHEYRLE 136
************.8899****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.12E-60 | 5 | 156 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.387 | 7 | 156 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.3E-29 | 8 | 135 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 358 aa Download sequence Send to blast |
MNSFSHVPPG FRFHPTDEEL VDYYLRKKVT SRRIDLDVIK DVDLYKIEPW DLQELCRIGT 60 EEQNEWYFFS HKDKKYPTGT RTNRATAAGF WKATGRDKGI YSKHDLIGMR KTLVFYKGRA 120 PNGQKSDWIM HEYRLETDPN GAPQAKGWVV CRVFKKKIAT MRRESEHESP IWYDDHVSFM 180 PEIDSPKQHS QSSLTYHYPN YPCKKELDNL HYQVPPDHHQ QQQFLQLPLL ETPKFLPAPP 240 SINCSSMPVF GINFNNHGSS LPQSSVLTRE HVHPMYGNNN NSTNNTNEQL ADQVTDWRVL 300 DKFVASQLSQ EEVISKENDY SNAGNTLHGH GNTNLNKQET ATENTSTASS CSQIDLWK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 3e-49 | 6 | 157 | 14 | 169 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:16103214, PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
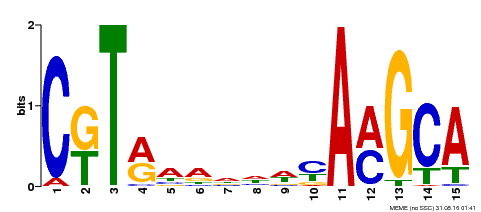
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00136 | DAP | Transfer from AT1G12260 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975515 | 3e-96 | HG975515.1 Solanum lycopersicum chromosome ch03, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019258894.1 | 0.0 | PREDICTED: NAC domain-containing protein 7-like | ||||
| Swissprot | Q9FWX2 | 1e-142 | NAC7_ARATH; NAC domain-containing protein 7 | ||||
| TrEMBL | A0A314LF71 | 0.0 | A0A314LF71_NICAT; Nac domain-containing protein 7 | ||||
| STRING | EOY22909 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA134 | 24 | 297 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G12260.1 | 1e-120 | NAC 007 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



