 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00351g00108.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 507aa MW: 55650.1 Da PI: 6.8544 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 41.5 | 2.4e-13 | 334 | 373 | 11 | 55 |
HHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 11 rRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
rRRdriN+++ L+ellP++ +K +Ka++L +A+eY+ksLq
Peaxi162Scf00351g00108.1 334 RRRDRINEKMKALQELLPHS-----TKTDKASMLDEAIEYLKSLQ 373
8******************9.....9******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-15 | 314 | 379 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-15 | 314 | 382 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.45E-16 | 319 | 377 | No hit | No description |
| SMART | SM00353 | 4.7E-13 | 322 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 14.037 | 323 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.2E-11 | 334 | 373 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 507 aa Download sequence Send to blast |
MMLLILLLGT SFDHELVELL WRNGEVVLHS QTHKKQGYDP NDCRQVNKHD QSTIRTSVSC 60 GNQANLIQDD ETISWLNCPI DDPFDKEFCS PFLSEISTNP IEADKITRQS EDNKVFKFDP 120 FEVNHVLPHS HHSGFDPNPM PPPKYHNSGS AQQQHVGGLQ KGVNFPTPVR SSNVQLGGKE 180 VSNMMLHDVK EGSVMTVGSS HCGSNQVAND IDTSRFSSST NRGLSTAMIT DYTGKVSPQS 240 DTMDRDTFEP ANTSSSGGSG SSYARTCNQS TATNSHSHKR KSREGEEPEC QSKSGDLDSA 300 GGNKSAQKSG TARRSRAAEV HNLSERVEFV GLTRRRDRIN EKMKALQELL PHSTKTDKAS 360 MLDEAIEYLK SLQMQLQMMW IGGGMASMMF PGVQHYMSRM GIGMGPPTVS SIHNAMNLAR 420 LPLVDPAIPL TQAAPNQAAM CQTSILNAVN YQRQLQNPNF SDQYASYMGF HPLQGASQPI 480 NIFSLGSHSA QQSQQIPPPT NSNAPTT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
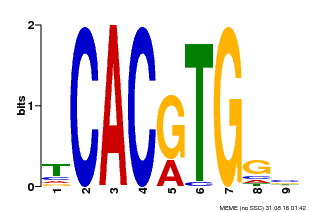
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016580439.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| TrEMBL | A0A1U8HIU3 | 0.0 | A0A1U8HIU3_CAPAN; Uncharacterized protein | ||||
| TrEMBL | A0A2G2Z1M5 | 0.0 | A0A2G2Z1M5_CAPAN; transcription factor PIF4 | ||||
| STRING | Solyc07g043580.2.1 | 0.0 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA10465 | 22 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 4e-55 | phytochrome interacting factor 4 | ||||



