 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peaxi162Scf00377g00029.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 421aa MW: 45142.9 Da PI: 8.6042 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 36.5 | 8.9e-12 | 248 | 287 | 11 | 55 |
HHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 11 rRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+RRd+iN+++ +L++l+P++ K +Ka++L ++++Y+k+Lq
Peaxi162Scf00377g00029.1 248 KRRDKINQRMKTLQKLVPNS-----NKTDKASMLDEVIDYLKQLQ 287
8******************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.75E-15 | 228 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.1E-15 | 228 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.08E-14 | 235 | 291 | No hit | No description |
| SMART | SM00353 | 1.8E-12 | 236 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 14.811 | 237 | 286 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.3E-9 | 248 | 287 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 421 aa Download sequence Send to blast |
MHGLGPRTRV PNNKPISNYG DTLESIVNQA TRCNIPHHHL KSAVDGNGHG GDEVVPWFNN 60 HPAVAGAPPV TGGSATLAMD ALVPCSKNPA TFNHHHRQSS SVHVPGVDGS THVGSCSGAT 120 ADWMTAPRVS NMEPSSRGGG ADMSVSGSAT CSRQLTIDTF DREMGVTTTG MFTSTSMGSP 180 ENTSSDKQFT NRTGDDHDSV CHSRAQREAG DEEDNKKGSK NSSSSTKRKR AAAIHNQSER 240 VSIYVSLKRR DKINQRMKTL QKLVPNSNKT DKASMLDEVI DYLKQLQAQV HMMSRMNMSP 300 AMVLAMQQQQ LQMSMMGMGM GMGMGMGVAG VVDMNTLSRP NITGLPPFLH PTAAAAFMQP 360 MASWPDNSSD RLTAAANVIP DPLAALLACQ SQPMTMDAYS RMAALYQQFQ QPATSSVPKN 420 G |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
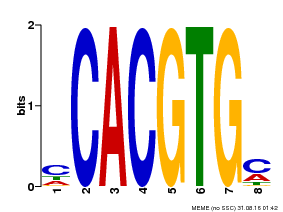
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009586991.1 | 0.0 | PREDICTED: transcription factor UNE10 isoform X1 | ||||
| Refseq | XP_009586993.1 | 0.0 | PREDICTED: transcription factor UNE10 isoform X3 | ||||
| TrEMBL | A0A1S4ALX7 | 0.0 | A0A1S4ALX7_TOBAC; transcription factor UNE10-like | ||||
| STRING | XP_009586991.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4367 | 21 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 1e-67 | bHLH family protein | ||||



