 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00087g11014.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 403aa MW: 44170.9 Da PI: 6.6201 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 27.3 | 7.9e-09 | 209 | 267 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i+eLe+k ++L++ ++L +l l+ +++e+
Peinf101Scf00087g11014.1 209 KRAKRIWANRQSAARSKERKMRYIAELERKLQALQTDATTLSAQLTLLQRDTNGMTAEN 267
9********************************************99998877777665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.7E-14 | 205 | 269 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.277 | 207 | 270 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 7.7E-9 | 209 | 262 | No hit | No description |
| SuperFamily | SSF57959 | 1.21E-9 | 209 | 259 | No hit | No description |
| Pfam | PF00170 | 1.8E-7 | 209 | 263 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 2.10E-23 | 210 | 259 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 403 aa Download sequence Send to blast |
MDKDKSSNHG GGLLPPSGSG RYSLFSPPGV KSEPNLPPLG PPGNSSDQSG HFDSSRFSHD 60 ISRMPDNPPK NLGHRRAHSE ILTLPDDISF DSDLGVVGGL DGPSLSDETE EDFFSMYLDM 120 DKFNSSSATS TFQVGESSSS AAAASGTENV APVIREKPRI RHQHSQSMDG STTIKPEMLT 180 SGAEEPSPAD AKKSMSAAKL AELALVDPKR AKRIWANRQS AARSKERKMR YIAELERKLQ 240 ALQTDATTLS AQLTLLQRDT NGMTAENSEL KLRLQTMEQQ VHLQDALNDA LKEEIQHLKV 300 LTGQGGIVNG GAMLNFPTTF GGNQQFFSNN QAMHTLLTTQ QLQQLQIHSH KQQHQFQHHQ 360 LHQIQQQQQQ DQQLQQAGDI KLRSSFSSAQ NDNASDISSK GKD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
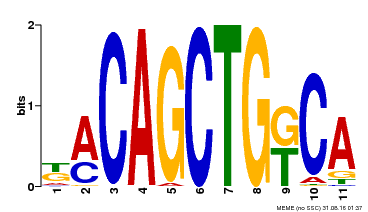
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027117495.1 | 0.0 | probable transcription factor PosF21 | ||||
| Refseq | XP_027117496.1 | 0.0 | probable transcription factor PosF21 | ||||
| Refseq | XP_027117497.1 | 0.0 | probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-149 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A068VB62 | 0.0 | A0A068VB62_COFCA; Uncharacterized protein | ||||
| STRING | XP_009605235.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3965 | 21 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-122 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



