 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00197g27013.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 269aa MW: 30542.8 Da PI: 5.3265 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 92.3 | 2.3e-29 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k ien nrqvtfskRr+g+lKKA+ELSvLCdaevaviifsstgkl+e+ss
Peinf101Scf00197g27013.1 9 KLIENVNNRQVTFSKRRAGLLKKANELSVLCDAEVAVIIFSSTGKLFEFSS 59
569**********************************************96 PP
| |||||||
| 2 | K-box | 51.4 | 4.6e-18 | 111 | 187 | 12 | 88 |
K-box 12 akaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelq 88
++ q+e++ Lk+e+++L+ q +llG+dL+ + l+eL+ Le+qL+++l i+++K+ell++q+e+++++ +el+
Peinf101Scf00197g27013.1 111 YVPKQEQKEMDILKDELSKLKMDQLRLLGKDLSGMGLNELRLLEHQLNEGLLAIKDRKEELLIQQLEQSRRQLEELR 187
556789*****************************************************************999875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 3.3E-41 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 32.001 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.12E-41 | 2 | 72 | No hit | No description |
| SuperFamily | SSF55455 | 1.03E-32 | 2 | 76 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.3E-28 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 4.5E-26 | 11 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.3E-28 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.3E-28 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 10.428 | 113 | 223 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 1.4E-12 | 114 | 186 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MGRGKIDIKL IENVNNRQVT FSKRRAGLLK KANELSVLCD AEVAVIIFSS TGKLFEFSST 60 SMKQTLSRYN RCLASTETSA IEKKLESRFL NFSFALSLNK DNEQPQPLQT YVPKQEQKEM 120 DILKDELSKL KMDQLRLLGK DLSGMGLNEL RLLEHQLNEG LLAIKDRKEE LLIQQLEQSR 180 RQLEELRGLF PLSTSLPPPY LEYHPLEQKY PILKEGEESL DSDTACEDGV DDEDSNTTLQ 240 LGLPIVGRKR KKPEQDSPSS NSENQVGSK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_B | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_C | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_D | 1e-21 | 1 | 69 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 247 | 251 | RKRKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
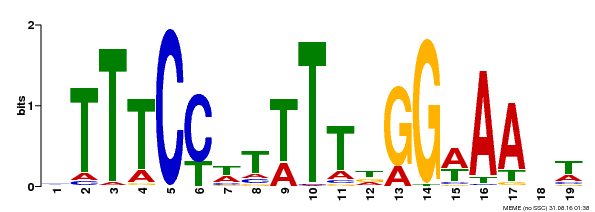
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY370526 | 1e-146 | AY370526.1 Petunia x hybrida MADS-box protein 9 (PMADS9) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009787834.1 | 1e-157 | PREDICTED: agamous-like MADS-box protein AGL15 | ||||
| Refseq | XP_016490205.1 | 1e-157 | PREDICTED: agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | Q6UGQ9 | 1e-175 | Q6UGQ9_PETHY; MADS-box protein 9 | ||||
| STRING | XP_009787834.1 | 1e-156 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA8775 | 22 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 3e-44 | AGAMOUS-like 15 | ||||



