 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00441g06019.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 403aa MW: 44252.8 Da PI: 7.081 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.8 | 2.2e-32 | 228 | 283 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Peinf101Scf00441g06019.1 228 KQRRCWSPELHRRFLQALQQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 283
79****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.875 | 225 | 285 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-29 | 226 | 286 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.14E-18 | 226 | 286 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.2E-28 | 228 | 283 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.1E-8 | 230 | 281 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0055062 | Biological Process | phosphate ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 403 aa Download sequence Send to blast |
MMINNNHDNH TEKMQRCQQY IDALEEERTK IQVFSRELPL CLELVTQAIE TYKQQLSGTT 60 TETYNLNAHE SDQCSEQTCS DHVPILEEFI PLKRTCSEED EEEEDDDNER VSHKSKTTNT 120 SNNSTSSKNN DKCCKKSDWL RSVQLWNQTS DPTPKEEVTP RQVSVVEVKK NGSGGAFHPF 180 KKEKSASTSA AAAAESPALT VVPAAAASST AESGGGGSKK EDKDSQRKQR RCWSPELHRR 240 FLQALQQLGG SHVATPKQIR ELMKVDGLTN DEVKSHLQKY RLHTRRPSPS IHNNNNQQPP 300 QFVVVGGIWV PPPEYAAMAA AAPAASGEAS GVAHSNGIYA PIATLPKGPF QDGTLKQRQH 360 ISNKPSRRSE ERGCGSHSDG GVHSNSPATS NSSTHTTTAS PAY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-13 | 228 | 281 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate homeostasis. Involved in the regulation of the developmental response of lateral roots, acquisition and/or mobilization of phosphate and expression of a subset of genes involved in phosphate sensing and signaling pathway. Is a target of the transcription factor PHR1. {ECO:0000269|PubMed:27016098}. | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
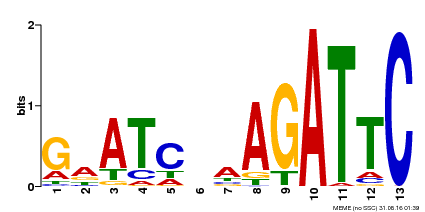
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00218 | DAP | Transfer from AT1G68670 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced under phosphate deprivation conditions. {ECO:0000269|PubMed:27016098}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016548806.1 | 0.0 | PREDICTED: transcription factor LUX | ||||
| Swissprot | Q8VZS3 | 5e-97 | HHO2_ARATH; Transcription factor HHO2 | ||||
| Swissprot | Q9FPE8 | 4e-97 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | A0A1U8EQX2 | 0.0 | A0A1U8EQX2_CAPAN; transcription factor LUX | ||||
| STRING | XP_009788853.1 | 1e-170 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5219 | 24 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68670.1 | 2e-80 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



