 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00898g01016.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 507aa MW: 55393.9 Da PI: 7.3242 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 107.1 | 8.8e-34 | 233 | 289 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cpvkkkvers d++v+ei+Y+g+Hnh+
Peinf101Scf00898g01016.1 233 DDGYNWRKYGQKQVKGSEYPRSYYKCTHTNCPVKKKVERSL-DGQVTEIIYKGQHNHQ 289
8****************************************.***************8 PP
| |||||||
| 2 | WRKY | 105.2 | 3.5e-33 | 395 | 453 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Cts+gC+v+k+ver+a+dpk+v++tYeg+H h+
Peinf101Scf00898g01016.1 395 LDDGYRWRKYGQKVVKGNPYPRSYYKCTSQGCNVRKHVERAASDPKAVITTYEGKHSHD 453
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.0E-30 | 226 | 291 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.065 | 227 | 291 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-26 | 230 | 291 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.7E-38 | 232 | 290 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.2E-26 | 233 | 289 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 4.0E-37 | 380 | 455 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-29 | 387 | 455 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.618 | 390 | 455 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.2E-39 | 395 | 454 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.6E-26 | 396 | 453 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 507 aa Download sequence Send to blast |
MADNEGSSSS SSSSRTPLLR PTITLPSRNS IETLFTGALS SGISPGPMTL VSSFFNDNDP 60 DSDCRSFSQL LAGAMSSPAG FPPPSTAGKD SGEFRFKENR PTGLVISQSS NTFTVPPGLS 120 PASLLDSPGF FSPGQGPFGM SHQQVLAQLT AQAAQPQSQM GHSQMHIQPD YPSSSAPPAP 180 SLPQFQSFTS NVAVNQPRAP PASDPTIVRE SSDISLSDQR SEPASIAVDK PADDGYNWRK 240 YGQKQVKGSE YPRSYYKCTH TNCPVKKKVE RSLDGQVTEI IYKGQHNHQP PQSSKRSKDN 300 GNPNGNYNLQ GPYESKEGEP SYSLSMKDQE SSQISGSSDS EEVGDAETRV DGRDSDERES 360 KRRAVEVQTS EAACSHRTVT EPRIIVQTTS EVDLLDDGYR WRKYGQKVVK GNPYPRSYYK 420 CTSQGCNVRK HVERAASDPK AVITTYEGKH SHDVPAARNS SHSTANSMSQ LRPQNSVVDK 480 PTAMRRTDFQ NNEQPITLLR FKEEQIT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-41 | 233 | 456 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-41 | 233 | 456 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
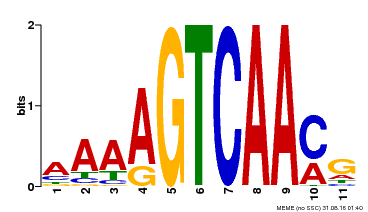
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009794188.1 | 0.0 | PREDICTED: probable WRKY transcription factor 4 | ||||
| Swissprot | Q9ZQ70 | 1e-170 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A1U7XTB8 | 0.0 | A0A1U7XTB8_NICSY; probable WRKY transcription factor 4 | ||||
| STRING | XP_009794188.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4107 | 24 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13960.1 | 1e-135 | WRKY DNA-binding protein 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



