 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00920g02019.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 750aa MW: 86085.7 Da PI: 6.3735 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 85.3 | 9.6e-27 | 94 | 196 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk.............tekerrtraetrtgCkaklkvkkekd 73
fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ e++ +ra +t+Cka+++vk++ d
Peinf101Scf00920g02019.1 94 FYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYEKSanrprsrqgnkqdPENATGRRACAKTDCKASMHVKRRPD 178
9***************************************999999899999888776555556699****************** PP
FAR1 74 gkwevtkleleHnHelap 91
gkw ++++e+eHnHel p
Peinf101Scf00920g02019.1 179 GKWLIHRFEKEHNHELLP 196
***************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.1E-24 | 94 | 196 | IPR004330 | FAR1 DNA binding domain |
| PROSITE profile | PS50966 | 9.409 | 472 | 508 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 5.0E-6 | 483 | 510 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 750 aa Download sequence Send to blast |
MDIDLRLPSR DHDKEEEEEE EEEQNGIINM LDNEEKLHSD EGMHGMLVVE EKMHAEDGED 60 MDNPITNMIE FKEDVNLEPL AGMEFESHGE AYAFYQEYAR SMGFNTAIQN SRRSKTSREF 120 IDAKFACSRY GTKREYEKSA NRPRSRQGNK QDPENATGRR ACAKTDCKAS MHVKRRPDGK 180 WLIHRFEKEH NHELLPAQAV SEQTRRMYAA MARQFAEYKN VVGLKSDAKG PFDKARNSAM 240 EGGDINVLLE FFIQMQALNS NFFYAVDVEA LPSSLHYFCM WHILGKVSEK LNHVIKQNEK 300 FMAKFEKCIY RSWTDEEFEK RWRKLVDRFD LREADLIRSL YEDRAKWAPT FMRDAFLAGM 360 STVQRSESVN SFIDKYVHKK TTVQEFVKQY ESILQDRYEE EAKADSDTWN KQPALKSPSP 420 FEKHVAGLYT HAVFKKFQGE VLGAVACIPK REKQDETTIT FRVQDFEKDQ EFIVTLNEVK 480 SEVSCICHLF ELKGYLCRHT LIVLQICGVS SIPSQYILKR WTKDAKSKYS VSDGSEDVQS 540 RVHRYNELCQ RAMKLSEEGS LSQESFSFAL RALDDAFGSC VTFNNSNKNM LEAGTSAAPG 600 LLCVEDDNQS RSMSKTNKKK NNFTKKRKVN SEADVMTVGA ADNLQQMDKL NSRPVTLDGY 660 FGAQQGVQGM VQLNLMAPTR DNYYGNQQTI QGLGQLNSIA PTHDGYYGAQ PAMHGLGQMD 720 FFRAPSFPYG IRDEPNVRSA QLHDESSRHP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
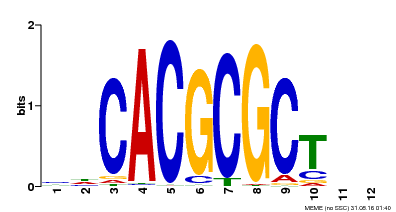
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT013715 | 0.0 | BT013715.1 Lycopersicon esculentum clone 132560F, mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019230455.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A1R3IYV4 | 0.0 | A0A1R3IYV4_9ROSI; Uncharacterized protein | ||||
| STRING | XP_009804946.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA496 | 21 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



