 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf00990g00013.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 722aa MW: 77611.7 Da PI: 6.3143 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.9 | 5.5e-16 | 443 | 489 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Peinf101Scf00990g00013.1 443 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 489
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.53E-17 | 435 | 493 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-20 | 436 | 497 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.7E-20 | 436 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 439 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.4E-13 | 443 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 445 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 722 aa Download sequence Send to blast |
MVRGKLESGQ PKISAATPFP ENDVVELVWE NGQIMMQGQS SSAKKSPTSN NLPSISPRDQ 60 DRYMGNSSTS KIGKFGVMDS MLNDMPSVPA GELDLLQEDE GVPWLGYSEQ QDYCAQLLPE 120 ISGMTENEPS RQNEFVLMDK RGSTDQVIGD SHSVPSHNAM NYEQRNTSKV SPSSRFSLLS 180 SLVPQKGHTS TLSLEPGVSD IFSSRTSYNP VSVFGDSSPS QASAGDVRSI TIRKQNMPES 240 SSGLLNFSHF SRPATFVKAA KLPNSAGGLN VSGSSTFEEK GIREGENVTN SNNNVSAASV 300 ENFLSSKKEN HYPTNVVSSQ LESRASGASF HDRSCPVEQS DVYTDCSKNN DNTPAQLTGA 360 KATKDIANGE RNVEHGVACS SVCSGSSAER GSSDQPHNLK RKTRDIEESG SRSEDIEEES 420 VGIKKACPTR GGTGSKRSRA AEVHNLSERR RRDRINEKMR ALQELIPNCN KADKASMLDE 480 AIEYLKTLQL QVQIMSMGAG MCVPPMMFPT GVQHMHATQM RHFSPMGLGM GMGMGMGFGM 540 GMLEMNGRSS GCPIFPMPPF QGAHFPSPPI PASTAYPGTA GSNPHAFAHP GQGLPMPIPR 600 ASRAPLAGQL STGSAVSLNV PRAGVPVDML SAPPILDSKN PVNKKNSKIE HNVEPSCPIN 660 QTCSQVQATN EVLEKSASLQ KNDQLPDVTD SAANNLTNQI DVPVSEAGEY RLDKTCDIQL 720 IQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 447 | 452 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
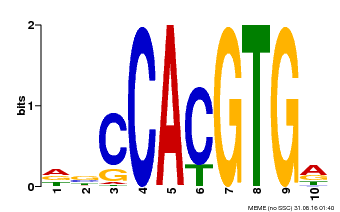
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006358519.1 | 0.0 | PREDICTED: transcription factor PIF3-like | ||||
| Refseq | XP_015169448.1 | 0.0 | PREDICTED: transcription factor PIF3-like | ||||
| TrEMBL | M1BK41 | 0.0 | M1BK41_SOLTU; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400047079 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 3e-47 | phytochrome interacting factor 3 | ||||



