 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf01166g07043.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 210aa MW: 24401.9 Da PI: 9.6538 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 94.1 | 6.2e-30 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien + rqvtfskRr g+lKKA+ELSvLCdaev++i+fs++gklye+ss
Peinf101Scf01166g07043.1 9 KRIENATSRQVTFSKRRSGLLKKAFELSVLCDAEVSLIVFSPKGKLYEFSS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 70.2 | 6.4e-24 | 79 | 171 | 6 | 98 |
K-box 6 gksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqee 90
+++++e+++e+l++ela + +++e L+ ++R+llG++L+s s+ eL+q+e+qLeksl++iR++Kn+l++eqi +l++ ek l ++
Peinf101Scf01166g07043.1 79 NRKTAEQTSEHLKEELATMTRKLELLEDSKRKLLGQGLDSSSFDELEQVEEQLEKSLSNIRARKNQLFREQIAHLKEEEKILMKQ 163
34478999***************************************************************************** PP
K-box 91 nkaLrkkl 98
n +Lrk+
Peinf101Scf01166g07043.1 164 NADLRKRC 171
*****986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 31.427 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.2E-40 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 1.83E-32 | 3 | 75 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.72E-41 | 3 | 72 | No hit | No description |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-30 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 3.7E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-30 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-30 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 5.5E-24 | 83 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.079 | 87 | 177 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000060 | Biological Process | protein import into nucleus, translocation | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010077 | Biological Process | maintenance of inflorescence meristem identity | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008134 | Molecular Function | transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 210 aa Download sequence Send to blast |
MVRGKTQLKR IENATSRQVT FSKRRSGLLK KAFELSVLCD AEVSLIVFSP KGKLYEFSSS 60 STNKTIERYQ KNDKYLAHNR KTAEQTSEHL KEELATMTRK LELLEDSKRK LLGQGLDSSS 120 FDELEQVEEQ LEKSLSNIRA RKNQLFREQI AHLKEEEKIL MKQNADLRKR CEVLPLPLTI 180 VPQEEEDPGR QRMEVETQLF IGLPQRQSSH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 2e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_B | 2e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_C | 2e-18 | 1 | 69 | 1 | 69 | MEF2C |
| 5f28_D | 2e-18 | 1 | 69 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor active in flowering time control. May control internode elongation and promote floral transition phase. May act upstream of the floral regulators MADS1, MADS14, MADS15 and MADS18 in the floral induction pathway. {ECO:0000269|PubMed:15144377, ECO:0000269|PubMed:17166135}. | |||||
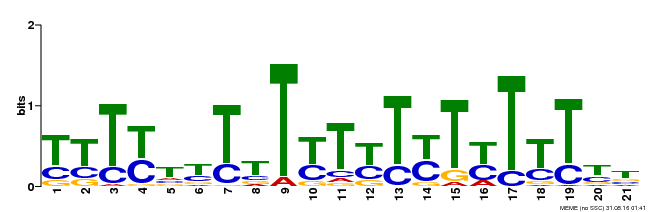
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00076 | ChIP-chip | Transfer from AT2G45660 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY370520 | 3e-73 | AY370520.1 Petunia x hybrida MADS-box protein 16 (PMADS16) gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009772098.1 | 1e-128 | PREDICTED: agamous-like MADS-box protein AGL19 | ||||
| Refseq | XP_016435497.1 | 1e-128 | PREDICTED: agamous-like MADS-box protein AGL19 | ||||
| Swissprot | Q9XJ60 | 4e-73 | MAD50_ORYSJ; MADS-box transcription factor 50 | ||||
| TrEMBL | A0A1S3X6K8 | 1e-127 | A0A1S3X6K8_TOBAC; agamous-like MADS-box protein AGL19 | ||||
| TrEMBL | A0A1U7W0H1 | 1e-127 | A0A1U7W0H1_NICSY; agamous-like MADS-box protein AGL19 | ||||
| STRING | XP_009772098.1 | 1e-127 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45660.1 | 8e-74 | AGAMOUS-like 20 | ||||



