 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Peinf101Scf01271g04026.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Petunioideae; Petunia; Petunia integrifolia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 417aa MW: 45560 Da PI: 10.1915 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 40.9 | 4.6e-13 | 339 | 387 | 5 | 53 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelk 53
+r rr++kNRe+A rsR+RK+a++ eLe +v++L++ N++L+k+ e+
Peinf101Scf01271g04026.1 339 RRRRRMIKNRESAARSRARKQAYTLELEAEVAKLKEINEELRKKQAEIV 387
689***************************************8755543 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.3E-11 | 335 | 400 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.875 | 337 | 389 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 6.7E-11 | 339 | 397 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.39E-9 | 339 | 387 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.9E-12 | 339 | 390 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 342 | 357 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 3.30E-22 | 344 | 393 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 417 aa Download sequence Send to blast |
MSSGTMGSYM NFKNFVDTPQ PESNGGKSMG NDNFPLARQS SIYSLTFDEL QSTYSGLGKD 60 FGSMNMEDLL KNIWTAEESQ ALASTTGGVD GNVPAGNLQR QGSLTLPRTL STKTVDEVWR 120 DFQKETVSTK DVSGTGRSNF GQRQSTLGEM TLEEFLVRAG VVREDMQETA NSNDIRFDNG 180 LSQQSNNNSL TIAFQQPTQN NGLLFNQIAA NNMLNVAGPT PSKQQSQHQQ PLFPKQTTMA 240 FASPMQLANN GQLASPGTRA PVVGMSSPSI NTSSMAQGGM MGMAGLHSGV SPAKVGSPGN 300 DFGATSNTDT SSLSPSPYAF TEGGRGRRSG SSLEKVVERR RRRMIKNRES AARSRARKQA 360 YTLELEAEVA KLKEINEELR KKQAEIVEKQ KNQLLEKMRM PWGNKLRCLR RTLTGPW |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
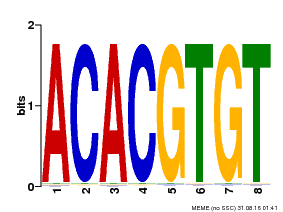
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016458525.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| Swissprot | Q9M7Q3 | 1e-105 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | A0A1S3Z283 | 0.0 | A0A1S3Z283_TOBAC; ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| STRING | XP_009774081.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA776 | 24 | 97 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34000.2 | 4e-88 | abscisic acid responsive elements-binding factor 3 | ||||



