 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.002G123700.1 | ||||||||
| Common Name | PHAVU_002G123700g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 658aa MW: 74578.9 Da PI: 6.3047 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 159.2 | 2.8e-49 | 55 | 197 | 2 | 140 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspes 92
++g+k+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGwvv++DGttyr+ + p+ +++gs+a +s+es
Phvul.002G123700.1 55 AKGKKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWVVDADGTTYRQ-CPPP---SHMGSFAAKSVES 141
689******************************************************************5.4444...69*********** PP
DUF822 93 slq.sslkssalaspvesysaspksssfpspssldsislasa.......asllpvl 140
+l+ sl++++++ +e++ ++++ ++ +sp+s+ds+ +a+ ++ +p+
Phvul.002G123700.1 142 QLSgGSLRNCSVKETIENQTSVLRIDECLSPASIDSVVIAERdsrnekyTNASPIN 197
***9***********************************98865555554444544 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.7E-50 | 56 | 202 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.05E-157 | 220 | 654 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.3E-171 | 223 | 653 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.6E-86 | 244 | 618 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 260 | 274 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 281 | 299 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 303 | 324 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 396 | 418 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 469 | 488 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 503 | 519 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 520 | 531 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 538 | 561 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-57 | 576 | 598 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 658 aa Download sequence Send to blast |
MKSINDDGST QDLDPQSDHS SDYLAHPNSN PHPHPHTRRP RGFAAAAAAT TNSVAKGKKE 60 REKEKERTKL RERHRRAITS RMLAGLRQYG NFPLPARADM NDVLAALARE AGWVVDADGT 120 TYRQCPPPSH MGSFAAKSVE SQLSGGSLRN CSVKETIENQ TSVLRIDECL SPASIDSVVI 180 AERDSRNEKY TNASPINTVD CLEADQLMQD IHSGVHENDF TGTPYVPVYV KLPAGIINKF 240 CQLIDPEGIR EELMHMKSLN VDGVVVDCWW GIVEGWSSQK YVWSGYRELF NIVRKFKLKL 300 QVVMAFHECG GNDSSDALIS LPQWVLDIGK DNQDIFFTDR EGRRNTECLS WGIDKERVLK 360 GRTGIEVYFD MMRSFRTEFD DLFAEGLIYA VEVGLGASGE LKYPSFSERM GWRYPGIGEF 420 QCYDKYLQHS LRKAAKLRGH SFWARGPDNA GHYNSMPHET GFFCERGDYD NYYGRFFLHW 480 YSQTLIDHAD NVLSLATLAF EETKIIVKVP AVYWWYKTPS HAAELTAGYH NPTNQDGYYP 540 VFEVLRKHAV TMKFVCLGFH LSSQEANESL IDPEGLSWQV LNSAWDRGLM AGGENALLCY 600 DREGYKRLVD TAKPRNDPDH RHFSFFVYQQ PSLLQANVCL SELDFFVKCM HGEMTDP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-127 | 225 | 615 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
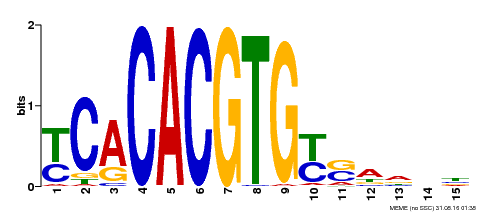
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.002G123700.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015043 | 1e-167 | AP015043.1 Vigna angularis var. angularis DNA, chromosome 10, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007158095.1 | 0.0 | hypothetical protein PHAVU_002G123700g | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | V7CIY1 | 0.0 | V7CIY1_PHAVU; Beta-amylase | ||||
| STRING | XP_007158095.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.002G123700.1 |
| Entrez Gene | 18637748 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



