 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.002G209300.1 | ||||||||
| Common Name | PHAVU_002G209300g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1087aa MW: 121650 Da PI: 5.8421 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 178.1 | 1.1e-55 | 22 | 136 | 4 | 118 |
CG-1 4 ekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
++rwl++ ei++iL n++ +++t e+++rp+sgsl+L++rk++ryfrkDG+ w+kkkdgktv+E+hekLKvg+v+vl+cyYah+een++f
Phvul.002G209300.1 22 AQNRWLRPAEICEILCNYRMFQITPEPPNRPPSGSLFLFDRKVLRYFRKDGHIWRKKKDGKTVKEAHEKLKVGSVDVLHCYYAHGEENESF 112
499**************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Le ++ +iv+vhyl+vk
Phvul.002G209300.1 113 QRRSYWMLEPDMMHIVFVHYLDVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.514 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.7E-80 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.9E-50 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 3.8E-4 | 499 | 588 | IPR013783 | Immunoglobulin-like fold |
| CDD | cd00102 | 0.00129 | 500 | 588 | No hit | No description |
| Pfam | PF01833 | 1.1E-5 | 501 | 586 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 1.01E-15 | 501 | 587 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 3.6E-18 | 685 | 795 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.64E-17 | 689 | 795 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.60E-13 | 700 | 786 | No hit | No description |
| SMART | SM00248 | 2800 | 701 | 730 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 20.208 | 701 | 809 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.576 | 701 | 733 | IPR002110 | Ankyrin repeat |
| Pfam | PF12796 | 2.8E-6 | 706 | 786 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0025 | 734 | 763 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.98 | 734 | 766 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.29 | 908 | 930 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 1.7E-7 | 909 | 959 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 8.114 | 909 | 938 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0022 | 911 | 929 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0071 | 931 | 953 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.285 | 932 | 956 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.3E-4 | 934 | 953 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1087 aa Download sequence Send to blast |
MAEPASYGLG PRLDLQQLQL EAQNRWLRPA EICEILCNYR MFQITPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHIWRKKKD GKTVKEAHEK LKVGSVDVLH CYYAHGEENE SFQRRSYWML 120 EPDMMHIVFV HYLDVKVNKT NVGGKTYSGE ATSDSQNGSS LSSGFPRNYG SVPSGSTDSM 180 SPTSTLTSLC EDADSEDIHQ ASSGLQSYHE SKSLGPMDKI DACSSSSYLT HPFSGDPAQF 240 PVPGAEYIPF VQGHKSRASD TAYTEGHRAH DIASWNNAME QSSGKHTATS LVSSTSIPTS 300 ASGNILEENN TVPGNLLGRK NALTEEERAS QPIHSNWQIP FEDDTIELPK WSLTQSLGLE 360 FGSDYGTSLL GDVTDTVGPE IVAEMFTFNG ELKEKSVHQN ISKQYTNTQS QPATKSNSEY 420 EVPGEASINY ALTMKRGLLD GEESLKKVDS FSRWITKEFA GVDDLHMQSS PGISWSTDDC 480 GDVIDDTSLN LSLSQDQLFS INDFSPKWAY AESEIEVLIV GTFLKSQPMV TACNWSCMFG 540 EVEVPAEVLA NGILCCQAPP HKIGRVPFYV TRANRFACSE VREFEYREGV DRNVDFADFF 600 NSATEMVLHL RLVGLLSLNS AHTSNQVFED DMEKRNLIFK LISLKEEEEY SCREETTVEM 660 DTTKHKLKEH MFHKQVKETL YSWLLRKVTE TGKGPRVLSE EGQGVLHLVA ALGYDWAIKP 720 IITAGVNINF RDASGWTALH WAAYCGRERT VAVLVSMGAD TKAVTDPCSE AREGRSPADL 780 ASSNGHKGLS GFLAESLLTS QLELLTMEEN KDGRKETSGM KAVQTVSERT ALPVLYGEVP 840 DAICLKDSLN AVRNATQAAD RIHQVYRMQS FQRKQLAQHD DDEFGLSDQQ ALSLLASRTN 900 KSGQGEGLAS AAAIQIQKKF RGWKKRKEFL IIRQRIVKIQ AHVRGHQVRK QYKPIIWSVG 960 ILEKVILRWR RKGSGLRGFR SDTVNKVVPD QPSESLKEDD YDFLKEGRKQ SEARFKKALS 1020 RVKSMVQYPE ARAQYRRVLN VVEDFRQTKG DNMNSMNSEE AVDGVEDLID IDMLLDDENF 1080 LPIAFD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
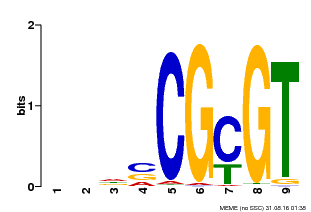
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.002G209300.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 0.0 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007159108.1 | 0.0 | hypothetical protein PHAVU_002G209300g | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | V7CLK6 | 0.0 | V7CLK6_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007159108.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7406 | 26 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.002G209300.1 |
| Entrez Gene | 18638623 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



