 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.003G229600.1 | ||||||||
| Common Name | PHAVU_003G229600g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 343aa MW: 38470.7 Da PI: 5.2628 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 169.2 | 1.4e-52 | 6 | 136 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkd 89
lppGfrFhPtdeelv +yLk+kveg ++el evi+ +d yk++Pw+Lp k + +++ ew+fF++rd+ky++g+r+nratk+gyWkatgkd
Phvul.003G229600.1 6 LPPGFRFHPTDEELVGYYLKRKVEGLEIEL-EVIPVIDFYKFDPWELPeKsFLPKRDMEWFFFCPRDRKYPNGSRTNRATKAGYWKATGKD 95
79****************************.99**************95344555788********************************* PP
NAM 90 kevlsk..kgelvglkktLvfykgrapkgektdWvmheyrl 128
++v+++ ++++g +ktLvfy grap g +tdWvmheyrl
Phvul.003G229600.1 96 RKVVCQsnPSNVTGYRKTLVFYAGRAPLGDRTDWVMHEYRL 136
****98666778***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 5.75E-60 | 4 | 159 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.748 | 6 | 159 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.6E-29 | 7 | 136 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008283 | Biological Process | cell proliferation | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 343 aa Download sequence Send to blast |
MGGATLPPGF RFHPTDEELV GYYLKRKVEG LEIELEVIPV IDFYKFDPWE LPEKSFLPKR 60 DMEWFFFCPR DRKYPNGSRT NRATKAGYWK ATGKDRKVVC QSNPSNVTGY RKTLVFYAGR 120 APLGDRTDWV MHEYRLCDDL GQPTPSFLGG FALCRVIKKN EKASATQGEN KGKRAGSSSI 180 NGTDTSVRFS GEQPFSNSGD SSSQASHLNN ESRYSSPISS PYNVPPMGEI NLASLETNPS 240 NFWISPDMIL DSSKDYPELQ HAVAEYFPGY DLPRMGTQFQ SLEHSETPSS FSYSNVNGEV 300 EFADTLSQIM SPYSGQWNSM DFYGKGGVPY EDHDQINSIS YP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-51 | 5 | 163 | 16 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-51 | 5 | 163 | 16 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-51 | 5 | 163 | 16 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-51 | 5 | 163 | 16 | 169 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 4e-51 | 5 | 163 | 16 | 169 | NAC domain-containing protein 19 |
| 4dul_B | 4e-51 | 5 | 163 | 16 | 169 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in tissue reunion of wounded inflorescence stems. Required for the division of pith cells in the reunion process, which is dependent on polar-transported auxin and the wound-inducible hormones ethylene and jasmonate (PubMed:21911380). Binds to the promoters of XTH19 and XTH20 to induce their expression via auxin signaling. XTH19 and XTH20 are involved in cell proliferation in the tissue reunion process of incised stems (PubMed:25182467). Involved in hypocotyl graft union formation. Required for the auxin- mediated promotion of vascular tissue proliferation during hypocotyl graft attachment (PubMed:27986917). {ECO:0000269|PubMed:21911380, ECO:0000269|PubMed:25182467, ECO:0000269|PubMed:27986917}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00439 | DAP | Transfer from AT4G17980 | Download |
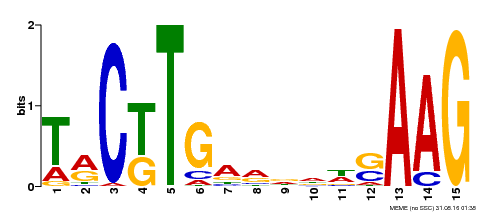
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.003G229600.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding in the flowering stem. {ECO:0000269|PubMed:21911380}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 1e-141 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007155764.1 | 0.0 | hypothetical protein PHAVU_003G229600g | ||||
| Swissprot | O49697 | 1e-104 | NAC71_ARATH; NAC domain-containing protein 71 | ||||
| TrEMBL | V7CEH0 | 0.0 | V7CEH0_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007155764.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6722 | 34 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17980.1 | 1e-106 | NAC domain containing protein 71 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.003G229600.1 |
| Entrez Gene | 18635777 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



