 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.003G248500.1 | ||||||||
| Common Name | PHAVU_003G248500g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 234aa MW: 24497 Da PI: 8.4938 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 121.1 | 4.1e-38 | 26 | 86 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++++l+cprC+stntkfCyynny+ sqPr+fCk+CrryWt+GG+lr++PvGgg+rkn k+s
Phvul.003G248500.1 26 EQENLPCPRCESTNTKFCYYNNYNYSQPRHFCKSCRRYWTHGGTLRDIPVGGGSRKNAKRS 86
57899****************************************************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF02701 | 6.4E-33 | 28 | 84 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.087 | 30 | 84 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 32 | 68 | IPR003851 | Zinc finger, Dof-type |
| ProDom | PD007478 | 2.0E-23 | 34 | 84 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0042545 | Biological Process | cell wall modification | ||||
| GO:0045787 | Biological Process | positive regulation of cell cycle | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 234 aa Download sequence Send to blast |
MPSSNSGESR RSAKPQSSAG APPPPEQENL PCPRCESTNT KFCYYNNYNY SQPRHFCKSC 60 RRYWTHGGTL RDIPVGGGSR KNAKRSRTHH TATSSSSSTM TSPQEHALPA LAPVPTTQGG 120 SMHFMDGDVN KQNNVNVCGS FTSLLNNTQG NNGFLALGGF GLGLGHGFED MGFGIGRSGW 180 GFPGMVDGAN IGGAVPSSGL STWQLESGEG GFVSGDCFSW PGLAISTPGN GLK* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Enhances the DNA binding of OBF transcription factors to OCS elements. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00402 | DAP | Transfer from AT3G50410 | Download |
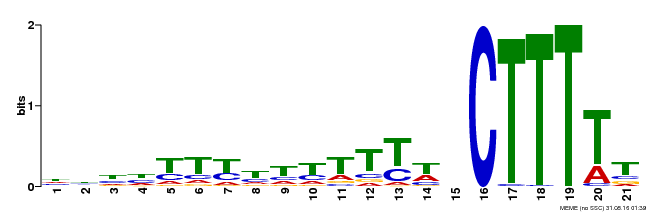
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.003G248500.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin and salicylic acid (SA). {ECO:0000269|PubMed:10758484}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015034 | 0.0 | AP015034.1 Vigna angularis var. angularis DNA, chromosome 1, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007155977.1 | 1e-171 | hypothetical protein PHAVU_003G248500g | ||||
| Swissprot | Q39088 | 4e-62 | DOF34_ARATH; Dof zinc finger protein DOF3.4 | ||||
| TrEMBL | V7CGC2 | 1e-170 | V7CGC2_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007155977.1 | 1e-171 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2801 | 31 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G50410.1 | 9e-53 | OBF binding protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.003G248500.1 |
| Entrez Gene | 18635971 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



