 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.007G206000.1 | ||||||||
| Common Name | PHAVU_007G206000g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 725aa MW: 77365.4 Da PI: 6.8158 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.7 | 3.3e-16 | 463 | 509 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Phvul.007G206000.1 463 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 509
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 2.6E-20 | 456 | 517 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-20 | 456 | 519 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 459 | 508 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.62E-17 | 462 | 513 | No hit | No description |
| Pfam | PF00010 | 1.1E-13 | 463 | 509 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 465 | 514 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 725 aa Download sequence Send to blast |
MPLYELYRLA REKLGTENNG TRASDQCSSP ENDFFELVWE NGQISSQGQS SRVRRSPSCR 60 SLPSHCLPSH SPKGRDKDVG CGASKMGKFR DLDSGLSEIS MSVPSREIDF CQDEDVLPWF 120 DYAAMDGSLQ HDYGSDFLAT STSFTLLDKK SHTNVALRDS HKTSEEHENA FKGSSAEQVE 180 STGPKSSTSQ LYPPSLHHCQ TSFVSSRSRA SDITENNNTS NANQDATYGE ITNFLSSSSD 240 FSSPKGKKQE PPLPGNGSSS TIMNFSHFAR PAAMVRANLQ NIGLKSGLSS ARPDSMEIKN 300 NGAAATSSNP PESIVIDSSG ECSKELTMHC QQVVDQSRSR TDLNTSLSKA VEQNAVLSKQ 360 SEPACNESST KIDQIADQVL GDSGAKGQTT TEKSMEAAVA SSSVCSGNGA DRGSDEPNQN 420 LKRKKKTPMN LSAIDAEEES AGVKKAAGGR GGAGSKRTRA AEVHNLSERR RRDRINEKMR 480 ALQELIPNCN KVDKASMLDE AIEYLKTLQL QVQIMSMGAG LYMPPMMLPA GMQHMHAPHM 540 APFTPVGVGM HMGYGMGYGV GMPDMNGGSL RFPMIQVPQM PQMQGSHIPL AHMSGPTALH 600 EMARSNLPGL GLPGQGHPMP MPRAPTLPFS GGPIMNSPAL GLHACGSSGL VETVDSASAS 660 GLKDQMPNVG PLLKQSTGGR DSTSQMPTHS EAAVVQFDQS ALVQSSGHTS EANANGAVNP 720 CPRR* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 467 | 472 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
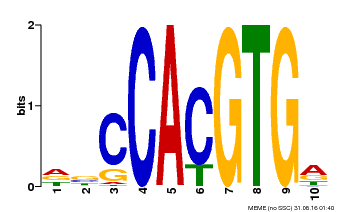
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.007G206000.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031132 | 0.0 | KT031132.1 Glycine max clone HN_WYQ_58 bHLH transcription factor (Glyma20g22291.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007145054.1 | 0.0 | hypothetical protein PHAVU_007G206000g | ||||
| TrEMBL | V7BJ54 | 0.0 | V7BJ54_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007145054.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5271 | 33 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-50 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.007G206000.1 |
| Entrez Gene | 18626525 |



