 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.010G103200.1 | ||||||||
| Common Name | PHAVU_010G103200g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 335aa MW: 37117.1 Da PI: 9.3702 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.4 | 1.6e-13 | 61 | 105 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
Phvul.010G103200.1 61 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 105
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.05E-16 | 55 | 111 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 56 | 110 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-6 | 59 | 111 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-18 | 59 | 108 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-11 | 60 | 108 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-11 | 61 | 105 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.09E-8 | 63 | 106 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MVSVNPNPAQ GFYFFDPSNM TLPGVNSLPP PPPPPQTPAA SVALEDPNKK IRKPYTITKS 60 RESWTEQEHD KFLEALQLFD RDWKKIEAFV GSKTVIQIRS HAQKYFLKVQ KNGTSEHVPP 120 PRPKRKATHP YPQKAPKITP TVSQVMGPLQ PSSAFIEPAY IYSPDSSSVL GTPLTNMPLS 180 SWNYNAKPPV NVPQVTRDNM GLTGAAQTVP LNCCYSSSNE NTPPTWSSSK KNHQGGQGKP 240 IKVMPDFAQV YSFLGSVFDP NSTNNRQKLR QMDPINVETV LLLMRNLSVN LMSPEFEDHK 300 KLLSSYDIDS DESKLDNICS KSLTDKSESA IMSA* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00423 | DAP | Transfer from AT4G01280 | Download |
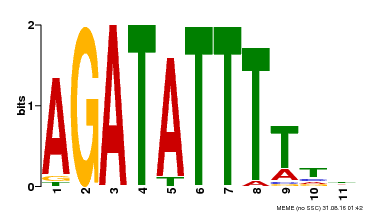
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.010G103200.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007135127.1 | 0.0 | hypothetical protein PHAVU_010G103200g | ||||
| Swissprot | C0SVG5 | 1e-108 | RVE5_ARATH; Protein REVEILLE 5 | ||||
| TrEMBL | V7ASD9 | 0.0 | V7ASD9_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007135127.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1119 | 33 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01280.1 | 1e-111 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.010G103200.1 |
| Entrez Gene | 18618019 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



