 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.011G071100.2 | ||||||||
| Common Name | PHAVU_011G071100g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 471aa MW: 51744.2 Da PI: 7.0286 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.5 | 5.9e-17 | 147 | 196 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Phvul.011G071100.2 147 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 196
78*******************......55************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 5.43E-16 | 147 | 205 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.3E-9 | 147 | 196 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.0E-32 | 148 | 210 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.27E-11 | 148 | 206 | No hit | No description |
| PROSITE profile | PS51032 | 17.662 | 148 | 204 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.4E-17 | 148 | 205 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.98E-13 | 239 | 284 | No hit | No description |
| SuperFamily | SSF54171 | 2.16E-6 | 239 | 283 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.1E-12 | 240 | 288 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 471 aa Download sequence Send to blast |
MLDLNLNAEW TDSARDGDSP LPLEKFPEGS RNQMAESGTS NSSVVNADGS SNGGCDEDSC 60 STRADDVYTF NFDILKVEGA SDVVTKELFP VSEGVATSSF SARKGFVDLA FDREGGNSEI 120 KMLQPQPQTQ TQQPAKKSRR GPRSRSSQYR GVTFYRRTGR WESHIWDCGK QVYLGGFDTA 180 HAAARAYDRA AIKFRGVNAD INFNLSDYED DFKQMKNLSK EEFVHILRRQ STGFSRGSSK 240 YRGVTLHKCG RWEARMGQFL GKKAYDKAAI KCNGREAVTN FEPSAYEGEI KSAAINEVGN 300 QNLDLNLGIA TPGPSPKENW EQLQFASVHY NTHGGISSMM ETNVNSGIGN PSLKRLVVSE 360 DCPSVWNGTC SSFFPNEEKT ERMNMDHSKG FPNWAWQTHA QVNATPVPTF SAAAASSGFS 420 ISATFPPTAT FPTRSMNCNP HQSLCFTSSS AHSSNASEYY YQVKPPQAPL * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
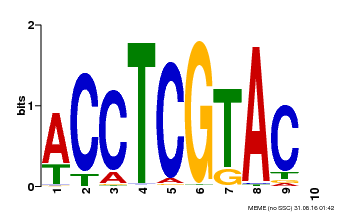
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.011G071100.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031086 | 0.0 | KT031086.1 Glycine max clone HN_CCL_205 AP2-EREBP transcription factor (Glyma12g07800.1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007132161.1 | 0.0 | hypothetical protein PHAVU_011G071100g | ||||
| Swissprot | Q9SK03 | 1e-125 | RAP27_ARATH; Ethylene-responsive transcription factor RAP2-7 | ||||
| TrEMBL | V7AFY0 | 0.0 | V7AFY0_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007132160.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.1 | 1e-122 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.011G071100.2 |
| Entrez Gene | 18615419 |



