 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Phvul.011G160400.1 | ||||||||
| Common Name | PHAVU_011G160400g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Phaseoleae; Phaseolus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 366aa MW: 40736.5 Da PI: 7.5978 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 177.5 | 3.6e-55 | 22 | 150 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdke 91
lppGfrFhPtdeel+++yL kkv +++++ ++i evd++k+ePw+Lp+k+k +ekewyf+s rd+ky+tg r+nrat++gyWkatgkd+e
Phvul.011G160400.1 22 LPPGFRFHPTDEELITYYLLKKVLDSSFTG-RAIVEVDLNKCEPWELPEKAKMGEKEWYFYSLRDRKYPTGLRTNRATEAGYWKATGKDRE 111
79*************************999.88***************99999************************************** PP
NAM 92 vlsk.kgelvglkktLvfykgrapkgektdWvmheyrle 129
++s+ + +lvg+kktLvfy+grapkgek++Wvmheyrle
Phvul.011G160400.1 112 IYSSkTCSLVGMKKTLVFYRGRAPKGEKSNWVMHEYRLE 150
**9745567****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.44E-62 | 18 | 174 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.387 | 22 | 174 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.9E-28 | 23 | 149 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010072 | Biological Process | primary shoot apical meristem specification | ||||
| GO:0010160 | Biological Process | formation of organ boundary | ||||
| GO:0010223 | Biological Process | secondary shoot formation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048504 | Biological Process | regulation of timing of organ formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MDSGYYNQRH HPHLDNNNEQ HLPPGFRFHP TDEELITYYL LKKVLDSSFT GRAIVEVDLN 60 KCEPWELPEK AKMGEKEWYF YSLRDRKYPT GLRTNRATEA GYWKATGKDR EIYSSKTCSL 120 VGMKKTLVFY RGRAPKGEKS NWVMHEYRLE GKFAYHYLSR SSKDEWVISR VFQKNNTGGA 180 STVSAAVATG GSKKTRISTS NTSSNMSLCP EPGSPSSIYL PPLLESSPYA ASSSAVATFN 240 DRENYSFESA AAAAAANQRE HVSCFSTLST DASAFNQLAP QPEPPLDPFS RFHRNNVGLS 300 AFPCLRSLHD NLNLPFFFPP MVHGGTDVAN FSAVANFPAP EDPRVVDGSS GMSIVPSELD 360 CMWGY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-50 | 15 | 177 | 10 | 168 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-50 | 15 | 177 | 10 | 168 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-50 | 15 | 177 | 10 | 168 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-50 | 15 | 177 | 10 | 168 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swm_B | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swm_C | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swm_D | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swp_A | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swp_B | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swp_C | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 3swp_D | 2e-50 | 15 | 177 | 13 | 171 | NAC domain-containing protein 19 |
| 4dul_A | 2e-50 | 15 | 177 | 10 | 168 | NAC domain-containing protein 19 |
| 4dul_B | 2e-50 | 15 | 177 | 10 | 168 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator of STM and KNAT6. Involved in molecular mechanisms regulating shoot apical meristem (SAM) formation during embryogenesis and organ separation. Required for the fusion of septa of gynoecia along the length of the ovaries. Activates the shoot formation in callus in a STM-dependent manner. Controls leaf margin development and required for leaf serration. Involved in axillary meristem initiation and separation of the meristem from the main stem. Regulates the phyllotaxy throughout the plant development. Seems to act as an inhibitor of cell division. {ECO:0000269|PubMed:10079219, ECO:0000269|PubMed:10750709, ECO:0000269|PubMed:12163400, ECO:0000269|PubMed:12492830, ECO:0000269|PubMed:12610213, ECO:0000269|PubMed:15202996, ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:15500463, ECO:0000269|PubMed:15723790, ECO:0000269|PubMed:16798887, ECO:0000269|PubMed:17098808, ECO:0000269|PubMed:17122068, ECO:0000269|PubMed:17251269, ECO:0000269|PubMed:17287247, ECO:0000269|PubMed:9212461}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00562 | DAP | Transfer from AT5G53950 | Download |
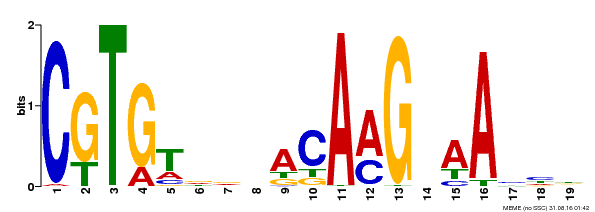
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Phvul.011G160400.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By BRM and SYD, at the chromatin level, and conferring a very specific spatial expression pattern. Precise spatial regulation by post-transcriptional repression directed by the microRNA miR164. {ECO:0000269|PubMed:15202996, ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:15723790, ECO:0000269|PubMed:16854978, ECO:0000269|PubMed:17251269, ECO:0000269|PubMed:17287247}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP015039 | 0.0 | AP015039.1 Vigna angularis var. angularis DNA, chromosome 6, almost complete sequence, cultivar: Shumari. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007133204.1 | 0.0 | hypothetical protein PHAVU_011G160400g | ||||
| Swissprot | O04017 | 1e-116 | NAC98_ARATH; Protein CUP-SHAPED COTYLEDON 2 | ||||
| TrEMBL | V7AK37 | 0.0 | V7AK37_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007133204.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3689 | 34 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G53950.1 | 1e-115 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Phvul.011G160400.1 |
| Entrez Gene | 18616353 |



