 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.001G361600.1 | ||||||||
| Common Name | POPTR_0001s37260g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 559aa MW: 60508.7 Da PI: 7.0081 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.1 | 6.6e-32 | 213 | 270 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
dDgynWrKYGqK+vkgsefprsYY+Ct+++C+vkk ers+ d++++ei+Y+g+H+h+k
Potri.001G361600.1 213 DDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERSH-DGQITEIIYKGTHDHPK 270
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.3 | 1.6e-33 | 378 | 436 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+v+g+++prsYY+Ct agCpv+k+ver+++dpk+v++tYeg+Hnh+
Potri.001G361600.1 378 LDDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHD 436
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 22.871 | 207 | 271 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.45E-25 | 210 | 271 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.0E-26 | 211 | 271 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.1E-34 | 212 | 270 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-24 | 213 | 269 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-37 | 363 | 438 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-29 | 370 | 438 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.825 | 373 | 438 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-39 | 378 | 437 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-26 | 379 | 436 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 559 aa Download sequence Send to blast |
MDNNTSRSEV SDCGDPTRPE SGGGGARYKL MSPAKLPISR SACITIPPGL SPTSFLESPV 60 LLSNVKAEPS PTTGTFTKPR TALGSLSSTP YSATTVSSTA CGERKSDYFE FRPYARSNMV 120 SADINHQRST QCAQVQSQCH SQSFASPPLV KGEMEVSTNE LSLSASLHMV TSVASAPAEV 180 DSDELNQTGL SSSGLQASQS DHRAGTAPSM SSDDGYNWRK YGQKHVKGSE FPRSYYKCTH 240 PNCEVKKLFE RSHDGQITEI IYKGTHDHPK PQPSRRYASG SVLSMQEDRF DKSSSLPNQG 300 DKSPGAYGQV PHAIEPNGAL ELSTGANDDT GEGAEDDDDP FSKRSRRLDA GGFDVTPVVK 360 PIREPRVVVQ TLSEVDILDD GYRWRKYGQK VVRGNPNPRS YYKCTNAGCP VRKHVERASH 420 DPKAVITTYE GKHNHDVPTA RTNSHDMAGP SAVNGPSRIR PDENETISLD LGVGISSTTE 480 NQSSDQQQAL HAELIKHENQ ASGSSFRVVH ATPITAYYGV LNGGMNQYGS RQIPGESRSI 540 EIPPYPYPQN MGRLLTGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 213 | 438 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 213 | 438 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. {ECO:0000250|UniProtKB:Q9SI37}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
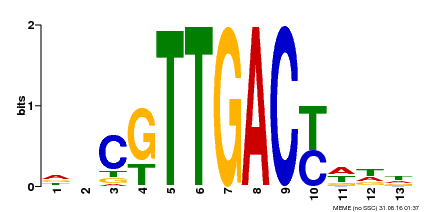
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.001G361600.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002298853.1 | 0.0 | probable WRKY transcription factor 20 isoform X1 | ||||
| Swissprot | Q93WV0 | 0.0 | WRK20_ARATH; Probable WRKY transcription factor 20 | ||||
| TrEMBL | B9GJX1 | 0.0 | B9GJX1_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0001s37260.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3946 | 34 | 63 | Representative plant | OGRP14 | 17 | 875 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 0.0 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.001G361600.1 |
| Entrez Gene | 7485085 |



