 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.003G097600.3 | ||||||||
| Common Name | POPTR_0003s09630g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 278aa MW: 29426 Da PI: 7.0814 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 70.5 | 2.5e-22 | 180 | 242 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr+rrkq+NRe+ArrsR+RK+ae++eL++++++L++eN L++e++++k e+++l +e+
Potri.003G097600.3 180 ERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLKEENANLRSEVNRIKSEYEQLLAEN 242
89*********************************************************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF16596 | 3.8E-57 | 12 | 147 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.2E-17 | 173 | 238 | No hit | No description |
| SMART | SM00338 | 6.1E-21 | 180 | 244 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.5E-20 | 180 | 242 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.242 | 182 | 245 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.25E-11 | 183 | 239 | No hit | No description |
| CDD | cd14702 | 4.20E-20 | 185 | 235 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 187 | 202 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 278 aa Download sequence Send to blast |
MEAEGRPSDA KEKLPIKRSK GSLGSLNMIT GKNNEHGRTT GASANGAYSK SAESGSEGTS 60 EGSDADSQSD SQMKSGGRQD SLEETSQNGG SAHAAQNGGQ GASTIMNQTM GVLPISAASA 120 PGVIPGPTTN LNIGMDYWGA PVASSVPAIR GKVPSTPVAG GIATAGSRDG VQSQHWLQDE 180 RELKRQRRKQ SNRESARRSR LRKQAECDEL AQRAEVLKEE NANLRSEVNR IKSEYEQLLA 240 ENASLKERLG EVHGQEDSRA GRNDQHTSND TQQTGQT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 196 | 202 | RRSRLRK |
| 2 | 196 | 203 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds to the G-box motif (5'-CACGTG-3') and other cis-acting elements with 5'-ACGT-3' core, such as Hex, C-box and as-1 motifs. Possesses high binding affinity to G-box, much lower affinity to Hex and C-box, and little affinity to as-1 element (PubMed:18315949). G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control (Probable). Binds to the G-box motif 5'-CACGTG-3' of LHCB2.4 (At3g27690) promoter. May act as transcriptional repressor in light-regulated expression of LHCB2.4. Binds DNA as monomer. DNA-binding activity is redox-dependent (PubMed:22718771). {ECO:0000269|PubMed:18315949, ECO:0000269|PubMed:22718771, ECO:0000305|PubMed:18315949}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
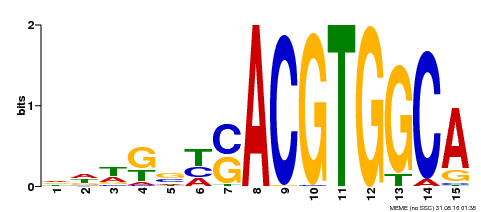
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.003G097600.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002304358.1 | 0.0 | bZIP transcription factor 16 | ||||
| Swissprot | Q501B2 | 1e-103 | BZP16_ARATH; bZIP transcription factor 16 | ||||
| TrEMBL | B9GZK1 | 0.0 | B9GZK1_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0003s09630.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G35530.1 | 2e-94 | basic region/leucine zipper transcription factor 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.003G097600.3 |
| Entrez Gene | 7491882 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



