 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.003G106500.3 | ||||||||
| Common Name | POPTR_0003s10570g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 676aa MW: 76390.7 Da PI: 6.0937 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 143.2 | 2.3e-44 | 73 | 197 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspes 92
g+g+++++kE+E++k+RER+RRai++++++GLR++Gn++lp+raD+n+Vl+AL+reAGw+ve DGttyr++ p ++gs+ + +es
Potri.003G106500.3 73 GKGKREREKEKERTKLRERHRRAITSRMLTGLRQYGNFPLPARADMNDVLAALAREAGWTVETDGTTYRQSPPPS----HTGSFGVRPVES 159
899******************************************************************776555....889998888888 PP
DUF822 93 slqsslkssalaspvesysaspksssfpspssldsislasa..a 134
+l k++a++ e + ++++ ++++sp slds+ +++ +
Potri.003G106500.3 160 PLL---KNCAVK---ECQPSVLRIDESLSPGSLDSMVISERenS 197
866...666655...45667788999999999999999877441 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.1E-44 | 73 | 210 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.56E-157 | 233 | 669 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.1E-171 | 235 | 668 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 1.1E-84 | 262 | 633 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 274 | 288 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 295 | 313 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 317 | 338 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 410 | 432 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 483 | 502 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 517 | 533 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 534 | 545 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 552 | 575 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-54 | 590 | 612 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 676 aa Download sequence Send to blast |
MNTNNSNQDL LINPQQTQIP QPDPYSHLPH PVQPGPSPHP HPQTRRPRGF AATAAAAASA 60 DNTSAVSSPN ASGKGKRERE KEKERTKLRE RHRRAITSRM LTGLRQYGNF PLPARADMND 120 VLAALAREAG WTVETDGTTY RQSPPPSHTG SFGVRPVESP LLKNCAVKEC QPSVLRIDES 180 LSPGSLDSMV ISERENSRNE KYTSTSPINS VIECLDADQL IQDVHSGMHQ NDFTENSYVP 240 VYVMLAQNGF INNCCQLIDP QGVRQELSHM KSLDVDGVVV ECWWGVVEAW SPQKYAWSGY 300 RELFNIIQEF KLKLQVVMAF HEYGGTDSGD VLISLPQWVL EIGKDNQDIF FTDREGRRNT 360 ECLSWGIDKE RVLKGRTGIE VYFDFMRSFR TEFNDLFTEG LITAIEIGLG PSGELKYPSF 420 SERIGWRYPG IGEFQCYDKY SQQNLRKAAK LRGHSFWARG PDNAGQYNSR PHETGFFCER 480 GDYDSYFGRF FLHWYSQSLI DHADNVLSLA SFAFEDTKII IKVPAVYWWY RTASHAAELT 540 AGYYNPTNQD GYSPVFEVLK KHSVIMKFVC SGLPLSGFEN DEALVDPEGL SWQILNSAWD 600 RGLTVAGVNM LACYDREGYR RVVEMAKPRN DPDHHHFSFF VYQQPSALAQ GTICFPELDY 660 FIKCMHGEIT GDLVS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-122 | 237 | 629 | 10 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
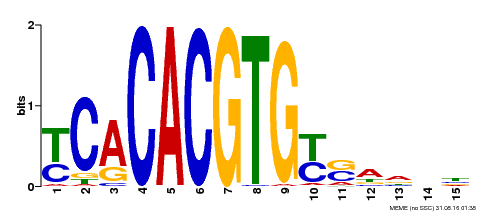
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.003G106500.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024453738.1 | 0.0 | beta-amylase 8 isoform X1 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A2K2B4Z4 | 0.0 | A0A2K2B4Z4_POPTR; Beta-amylase | ||||
| STRING | POPTR_0003s10570.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.003G106500.3 |
| Entrez Gene | 7491925 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



