 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.003G143000.2 | ||||||||
| Common Name | POPTR_0003s14300g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 954aa MW: 104676 Da PI: 5.4899 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 91.5 | 6.3e-29 | 574 | 625 | 1 | 52 |
RWP-RK 1 aekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
aek+isle+l++yF++++kdAAk Lgvc+T++KriCRq+GI+RWP+Rki+++
Potri.003G143000.2 574 AEKTISLEVLQQYFAGSLKDAAKRLGVCPTTMKRICRQHGISRWPSRKINKV 625
589***********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.302 | 564 | 645 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 6.0E-26 | 577 | 625 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 3.6E-22 | 850 | 941 | No hit | No description |
| SMART | SM00666 | 5.5E-25 | 857 | 939 | IPR000270 | PB1 domain |
| PROSITE profile | PS51745 | 24.619 | 857 | 939 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 6.6E-26 | 857 | 938 | No hit | No description |
| CDD | cd06407 | 1.45E-34 | 858 | 938 | No hit | No description |
| Pfam | PF00564 | 6.8E-18 | 858 | 938 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 954 aa Download sequence Send to blast |
MQKPIEGRGV GGGGGEPEGM KSMELDLDLD NSWPLDQISF MSSNPMSPFL ISTSTEQPCS 60 PLWAFSDAVD DRLAATASGQ ASPAFAAAAA PRLSDYPILL TCNPNLITES QGENDDNSKL 120 PSPFLGLMPI DNPDGYCMIK ERMTQALRYF KESTEQHVLA QVWAPVKNGG QHVLTTSGQP 180 FVLDPHSNGL HQYRMVSLMY MFSVDGESDR ELGLPGRVFR QKSPEWTPNV QYYSSKEYSR 240 LDHALRYNVR GTLALPVFEP SGQSCVGVLE LIMNSQKINY APEVDKVCKA LEAVNLKSSE 300 ILDPPSIQIC NEGRQNALSE ILEILTMVCE THKLPLAQTW VPCIHRSVLT YGGGLKKSCT 360 SFDGNCNGQV CMSTTDVAFY VVDARMWGFR EACLEHHLQK GQGVAGRAFL SQNSCFCPDI 420 TQFCKTEYPL VHYARMFGLT SCFAIFLRSS YTGDDDYILE FFLPPSITDS HEQKTFLGSI 480 LATMKQDFQS LKVASGMDLE EEGFVEMIEA TTNGRLECIQ IPQPTKSPPG DNMLPNEGHI 540 EQIDSEKNKL MFDLDVIKNG GRTKKPTERK RGKAEKTISL EVLQQYFAGS LKDAAKRLGV 600 CPTTMKRICR QHGISRWPSR KINKVNRSLS KLKWVIESVQ GTEGTFDLTP LTTSPLHVAD 660 GTISWPSNLN GSNQQTSPNS KPPEYHGNRN GSPTCRKPGS DGQAGSNRSK KRSGSRDGSA 720 GTPTSHDSCQ GSPENESAPV KDPSVSPVHE RCIKAGGSPG LALQQTKEQN LSSAYSIPDA 780 LVATEAHEPF GGMLIEDAGS SKDLRNLCPA VAEAIVDERV PESSWTDPPC FNMLPTQMFA 840 APLHAIPQAT PRQEMKSVTI KATYREDVIR FRISLSSGIV ELKEEVAKRL KLEVGTFDIK 900 YLDDDQEWVL IACDADLLEC MDVSRSSSSN IIRLSVHDAN ANLGSSCEST GEL* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, flowers and siliques. Detected in root hairs, emerging secondary roots, vascular tissues, leaf parenchyma cells and stomata. {ECO:0000269|PubMed:18826430}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in regulation of nitrate assimilation and in transduction of the nitrate signal. {ECO:0000269|PubMed:18826430}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
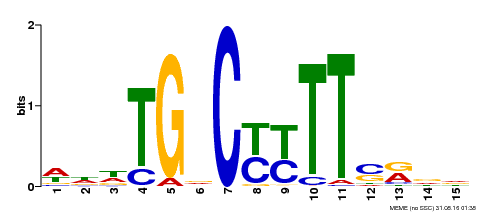
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.003G143000.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by the N source or by nitrate. {ECO:0000269|PubMed:18826430}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024453644.1 | 0.0 | protein NLP7 | ||||
| Swissprot | Q84TH9 | 0.0 | NLP7_ARATH; Protein NLP7 | ||||
| TrEMBL | A0A2K2B726 | 0.0 | A0A2K2B726_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0003s14300.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.003G143000.2 |
| Entrez Gene | 7497869 |



