 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.003G207200.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 449aa MW: 49944 Da PI: 7.3122 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.3 | 2.4e-12 | 382 | 427 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L++l+P+ +k + +++L +Av+YIk+Lq
Potri.003G207200.1 382 SIAERVRRTRISERMRKLQDLVPNM----DKQTNTSDMLDLAVDYIKDLQ 427
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.926 | 376 | 426 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.83E-15 | 379 | 439 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.01E-12 | 380 | 429 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.1E-14 | 381 | 437 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.4E-11 | 382 | 432 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.7E-9 | 382 | 427 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0001228 | Molecular Function | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 449 aa Download sequence Send to blast |
MDLLGSVSCY FCSASLFKLN QETGDSMESD LQHQHHFLHG HNQHQQIHQK QMNSGLTRYQ 60 SAPSSYFSSN LDRDFCEEFL NRPTSPETER IFARFLANSG GNTENIPGSN LCEIKQDSPV 120 KESVSQINQQ PQMMASMNNH SSDTRLHQHQ HQQHQHGNYS ASQGFYQSRS KPPLPDHNPG 180 SGMNHRSTNS TGLERMPSMK PSSGNNPNLV RHSSSPAGLF SNINIEFENG YAVLRDVGDL 240 GAGNRDTTYS AAGRPPSSSG IRSTIAEMGN KNMGENSPDS GGFGETPGNN YDYPIGSWDD 300 SAVMSTGSKR YLTDDDRTLS GLNSSETQQN EEAGNRPPML AHHLSLPKTS AEMSTIENFL 360 QFQDSVPCKI RAKRGCATHP RSIAERVRRT RISERMRKLQ DLVPNMDKQT NTSDMLDLAV 420 DYIKDLQRQF KALSENRARC TCLKKQQP* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
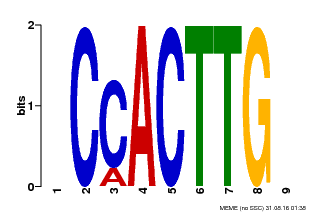
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.003G207200.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC216409 | 0.0 | AC216409.1 Populus trichocarpa clone POP024-J08, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002304761.1 | 0.0 | transcription factor bHLH122 isoform X1 | ||||
| TrEMBL | B9GW81 | 0.0 | B9GW81_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0003s22290.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4185 | 34 | 55 | Representative plant | OGRP511 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 2e-69 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.003G207200.1 |



