 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.004G140600.9 | ||||||||
| Common Name | POPTR_0004s14790g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 415aa MW: 45326.6 Da PI: 10.027 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.9 | 3.4e-11 | 336 | 383 | 5 | 52 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleel 52
+r+rr++kNRe+A rsR K+a +++Le++v++L++ N+ L+ + e+
Potri.004G140600.9 336 RRQRRMIKNRESAARSRTLKQAHTQKLEDEVAKLKELNEVLQRKQAEI 383
79************************************9998765555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.5E-6 | 332 | 398 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.933 | 334 | 383 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14707 | 3.17E-20 | 336 | 383 | No hit | No description |
| SuperFamily | SSF57959 | 1.25E-8 | 336 | 384 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 1.8E-11 | 336 | 383 | No hit | No description |
| Pfam | PF00170 | 1.5E-9 | 336 | 386 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MDSQWDYKNF GNAPPGQSSV TKPQENPFLI RQSSVYSLTF DEFQNTWGGG LRKDFGSMNM 60 EELLKNIWTA EETQAMTNTL GVGSEGSAPG GNLQRQGSLT LPRTLSQKTV DELWRDLIKE 120 TSGAAEDGSG SAGSNLPQRQ QTLGETTLEE FLVRAGVVRE DTQQIGRPDN SGFFGELSLL 180 NNNNDSSLAI GFQQPNGNNG LMGTWRMENN GNLVANQPPS LTLDAGGIRP TQQLPQSRQL 240 SQQQQLLFPK PAATVAFASP LHLSNNAQLA SPGVRRSVVG IADRSVNNGL AHSGGMGIVS 300 LATGGSNADT SSLSPVPYVF SRGRKASTAL EKVAERRQRR MIKNRESAAR SRTLKQAHTQ 360 KLEDEVAKLK ELNEVLQRKQ AEIIEMQQNQ FFETKKAQWG GKRQCLRRTL TGPW* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, leaves, flowers and immatures siliques. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:11884679}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
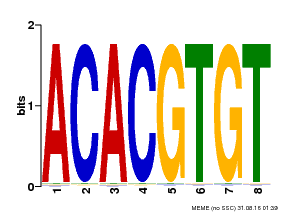
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.004G140600.9 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA) and cold. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF405964 | 0.0 | EF405964.1 Populus trichocarpa abscisic acid responsive elements-binding protein 2 (ABF2-1) mRNA, complete cds. | |||
| GenBank | EF405965 | 0.0 | EF405965.1 Populus trichocarpa abscisic acid responsive elements-binding protein 2 (ABF2-2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006384406.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 isoform X1 | ||||
| Refseq | XP_024455201.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 isoform X1 | ||||
| Refseq | XP_024455202.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 isoform X1 | ||||
| Refseq | XP_024455203.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 isoform X1 | ||||
| Refseq | XP_024455204.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 isoform X1 | ||||
| Swissprot | Q9M7Q2 | 1e-105 | AI5L7_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| TrEMBL | A3FM74 | 0.0 | A3FM74_POPTR; Abscisic acid responsive elements-binding protein 2 | ||||
| STRING | POPTR_0004s14790.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G19290.1 | 1e-104 | ABRE binding factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.004G140600.9 |
| Entrez Gene | 18097783 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



