 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.005G001800.1 | ||||||||
| Common Name | POPTR_0005s00360g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 743aa MW: 79784.4 Da PI: 6.2408 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.4e-16 | 470 | 516 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Potri.005G001800.1 470 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 516
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.01E-17 | 462 | 520 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-20 | 463 | 527 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.4E-20 | 463 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 466 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-13 | 470 | 516 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 472 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 743 aa Download sequence Send to blast |
MPLSELLYRM AKGKIDFSQE KDPSCSTDLS FVRPENDFGE LIWENGQIQS SRARKIQPCS 60 TLPCQNPKIR YKDIGNGTDI RTGKFGMMES TLNELPMSVP AVEMGVNQDD DMVPWLNYPL 120 DESPQHDYCS EFLPELSGVT VNGHSSQSNF PSFGKKSFSQ SVRDSRTVSV HNGLSLEQGD 180 VAKNSSAGDT EANRPRTSAS QLYLSSSEHC QTSFPYFRSR VSAKNGDSTS NAAHHVVSVD 240 SIRAPTSGGG FPSIKMQKQV PAQSTTNSSL MNFSHFARPA ALAKANLQNI GMRAGTGISN 300 MERTQNKDKG SIASSSNPAE CTPINSCSGL LKETSSHCLP VLMPPKVDAK PSEAKPAEGF 360 VPAELPEATI PEGDSKSDRN CRQNFCESAI KGVADVEKTK EPVVASSSVG SDNSVERASD 420 DPTENLKRKH RDTEESEGPS EDVEEESVGA KKQAPARAGN GSKRNRAAEV HNLSERRRRD 480 RINEKMRALQ ELIPNCNKVD KASMLDEAIE YLKTLQLQVQ IMSMGAGLYM PSMMLPPGMP 540 HMHAAHMGQF LPMGVGMGMR MGMGMGFGMS MPDVNGGSSG CPMYQVPPMH GPHFSGQPMS 600 GLSALHRMGG SNLQMFGLSG QGFPMSFPCA PLVPMSGGPP LKTNMEPNAC GVVGATDNLD 660 SATACSSHEA IQKINSQVMQ NNVVNSSMNQ TSSQCQATNE CFEQPALVQN NAQDSGVADN 720 RALKSVGGND NVPSSEAGEH LQ* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 474 | 479 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
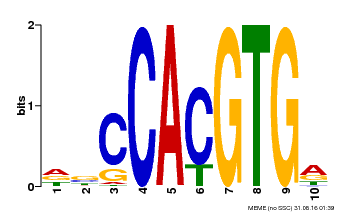
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.005G001800.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006382253.1 | 0.0 | transcription factor PIF3 isoform X1 | ||||
| TrEMBL | U5G9H3 | 0.0 | U5G9H3_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0005s00360.1 | 0.0 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5271 | 33 | 53 | Representative plant | OGRP258 | 16 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 2e-74 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.005G001800.1 |
| Entrez Gene | 18098446 |



