 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.005G170500.4 | ||||||||
| Common Name | POPTR_0005s16400g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 248aa MW: 28084.6 Da PI: 7.421 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 39.7 | 1.1e-12 | 74 | 115 | 2 | 43 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNk 43
+++k +rr+++NReAAr+sR+RKka++++Le +L + +
Potri.005G170500.4 74 PADKIQRRLAQNREAARKSRLRKKAYVQQLESSRMKLAQLEQ 115
57999**************************86555544443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.4E-9 | 70 | 121 | No hit | No description |
| SMART | SM00338 | 9.1E-7 | 73 | 133 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.5E-9 | 75 | 116 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.151 | 75 | 119 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14708 | 4.42E-22 | 77 | 129 | No hit | No description |
| SuperFamily | SSF57959 | 1.05E-7 | 77 | 119 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 80 | 95 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 3.1E-29 | 163 | 236 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 248 aa Download sequence Send to blast |
MSSTSTQIAA LRGMGIYEPF NQISSWAHAF RDDGSLNIGP STIVQVDAGL DDKSEHVSHE 60 SMEPYRSDQE AHKPADKIQR RLAQNREAAR KSRLRKKAYV QQLESSRMKL AQLEQELERA 120 RHQGAYLGSA SNSSHLGFSG TVNPGIAAFE MEYGHWVEEQ HKQISELRKA LQAHITDIEL 180 RILVENGLNH YNNLFRMKAD AAKADVFYLI SGKWRTSVER FFQWIGGFRP SELLNVSLIP 240 YNLLLED* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pth.11053 | 0.0 | bud| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
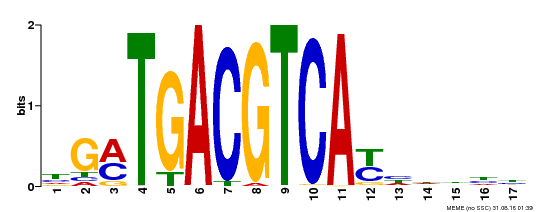
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.005G170500.4 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF145013 | 0.0 | EF145013.1 Populus trichocarpa clone WS0111_I20 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002306598.1 | 1e-175 | transcription factor TGA7 | ||||
| Swissprot | Q93ZE2 | 1e-101 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A385ZA14 | 1e-174 | A0A385ZA14_9ROSI; BZIP1 (Fragment) | ||||
| TrEMBL | B9H780 | 1e-174 | B9H780_POPTR; BZIP family protein | ||||
| STRING | POPTR_0005s16400.1 | 1e-175 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 1e-103 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.005G170500.4 |
| Entrez Gene | 7469115 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



