 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.005G221100.3 | ||||||||
| Common Name | POPTR_0005s24240g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 467aa MW: 51448.2 Da PI: 6.7192 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 36.1 | 1.2e-11 | 321 | 367 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ +I +Lq
Potri.005G221100.3 321 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITFITDLQ 367
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.3E-45 | 48 | 232 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.259 | 317 | 366 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.21E-14 | 320 | 371 | No hit | No description |
| SuperFamily | SSF47459 | 1.03E-17 | 320 | 388 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.1E-9 | 321 | 367 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.7E-16 | 321 | 387 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.8E-14 | 323 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 467 aa Download sequence Send to blast |
MGDKFWVNGE KGMVESVLGV EACEFLITSA SKNLLNDLVS PPVSLGVQQG LVQLVEGFNW 60 NYAIFWHASG LKTGGSILVW GDGICRDPKG QGIGDGSSSG DGKSEGAEKR KEVKKRVLQK 120 LHMCFNGPDD DNFAASVDEV SDVEMFYLTS MYFTFRCDST YGPGEAYQSG RSIWALGMPS 180 CLGHYQLRSV LARSAGFQTV VFLPVKSGVL ELGSVKSIPE QHDFVEKARS IFGASNTAQA 240 KAAPKIFGRE LSLGSSKSRS ISINFSPKVE DELIFTSEPY TMQAMSTDQD YPKDDLSPQG 300 DERKPRKRGR KPANGREEPL NHVEAERQRR EKLNQRFYAL RAVVPNISKM DKASLLGDAI 360 TFITDLQKKI RVLETERGVV NNNQKQLPVP EIDFQPRQDD AVVRASCPME SHPVSTIIET 420 FREHQITAQD CNVSVEGDKI VHTFSIRTQG GAADQLKEKL EAALSK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_B | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_E | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_F | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_G | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_I | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_M | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_N | 4e-28 | 315 | 382 | 2 | 69 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 302 | 310 | RKPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
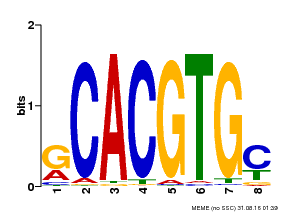
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.005G221100.3 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024457616.1 | 0.0 | transcription factor bHLH3 | ||||
| Swissprot | O23487 | 0.0 | BH003_ARATH; Transcription factor bHLH3 | ||||
| TrEMBL | A0A2K2AKL7 | 0.0 | A0A2K2AKL7_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0005s24240.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 1e-173 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.005G221100.3 |
| Entrez Gene | 7476894 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



