 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.007G093400.3 | ||||||||
| Common Name | POPTR_0007s05410g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1021aa MW: 115041 Da PI: 6.1005 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179.3 | 4.6e-56 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenp 92
l+ ++rwl++ ei++iL+n+++++++ e+++ p+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+v+vl+cyYah+e+n+
Potri.007G093400.3 20 LEAQHRWLRPAEICEILTNYQRFRIAPEPAHMPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKSGSVDVLHCYYAHGEDNE 110
5679*************************************************************************************** PP
CG-1 93 tfqrrcywlLeeelekivlvhylevk 118
+fqrr+ywlLeeel++ivlvhy+evk
Potri.007G093400.3 111 NFQRRSYWLLEEELSHIVLVHYREVK 136
***********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.041 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.2E-77 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.1E-49 | 21 | 134 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.9E-4 | 478 | 566 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 9.49E-16 | 480 | 565 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.9E-5 | 480 | 563 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 4.2E-15 | 655 | 771 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.1E-16 | 660 | 772 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 6.16E-11 | 666 | 769 | No hit | No description |
| PROSITE profile | PS50297 | 16.547 | 677 | 781 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF00023 | 5.0E-4 | 710 | 741 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.011 | 710 | 739 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.778 | 710 | 742 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3300 | 749 | 778 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.78E-8 | 863 | 917 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 2 | 866 | 888 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0042 | 869 | 887 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.206 | 869 | 896 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0039 | 889 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.597 | 890 | 914 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.1E-4 | 892 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1021 aa Download sequence Send to blast |
MADTKRYPLG NQLDIQQILL EAQHRWLRPA EICEILTNYQ RFRIAPEPAH MPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHER LKSGSVDVLH CYYAHGEDNE NFQRRSYWLL 120 EEELSHIVLV HYREVKGTRT NFNRIKEHEE CIPYSQETED TMPSSEMDTS VSSRFHPNGY 180 QVPTRTTDTT SMNSAQASEY EDAESVYNNQ ASSTFHSFLE VQKPAMERID TGSSVHYDHM 240 TFSSDYQGKL SAVPGMDVIS LAQVDKTKET NGTESACEPQ KVIDLPSWED VLENYARGTE 300 SVPFQTLLSQ DDTVGIIPKQ EDGILEKLLT NSFDKREDIG RYDLTARFPD QQLDSGNLIN 360 TLEPLCTQEN DLHIQNDIQI QPANADHGMT LEGKSMYSSS VKHHILDGSG TEGLKKLDSF 420 TRWMSKELGD VEPQVQSSSG SYWITAESEN GVDDSSNPSQ GNLDAYLLSP SLSQDQLFSI 480 IDFSPNWAYA GTEIKVLIMG RFLKGREAAE NCQWSIMFGE VEVPAEVIAD GVLRCNTPSH 540 KAGRIPFYVT CSNRVACSEV REFEYLSHTQ DITYYYSDSV TEDLNMRFGK LLSLSSVSPS 600 KYDSSSVDEI LSSKINSLLN EDNETWDQMF KLTSEEGFSS EKVKEQLVQK LLKEQLHVWL 660 LQKASEGGKG PSVLDEGGQG VLHFAAALGY DWALEPTIVA GVSVNFRDVN GWTALHWAAS 720 YGRERTVASL IHLGAAPGAL TDPTPKYPTS RTPADLASAN GHKGISGFLA ESALSAHLSS 780 LNLEKQDGKA AEFNDADLPS RLPLKDSLAA VCNATQAAAR IHQVFRVQSF QKKQLKEYGD 840 DKLGMSHERA LSLIAVKSQK AGQYDEPVHA AIRIQNKFRG WKGRKEFLII RQRIVKIQAH 900 VRGHQVRKNY RKIIWSVGIL DKIILRWRRK GSGLRGFKSE ALTDGSSMQV VQSKDDDDDF 960 LKEGRRQTEE RSQIALARVK SMHQHPEARE QYCRLRNVVA EIQEAKVTDY YFYSFSLLVA 1020 * |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, carpels, and siliques, but not in stigmas or other parts of the flower. {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:14581622}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
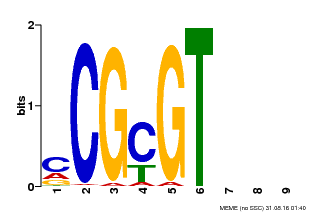
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.007G093400.3 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024461052.1 | 0.0 | calmodulin-binding transcription activator 3 isoform X2 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A2K1ZRQ8 | 0.0 | A0A2K1ZRQ8_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0007s05410.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.007G093400.3 |
| Entrez Gene | 7456909 |



