 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.009G064700.3 | ||||||||
| Common Name | POPTR_0009s06910g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 415aa MW: 45459.3 Da PI: 7.6177 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 33.1 | 9.9e-11 | 342 | 387 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L+el P+ +k + a++L +Ave+Ik+Lq
Potri.009G064700.3 342 SIAERVRRTRISERMRKLQELFPNM----DKQTNTADMLDLAVEHIKDLQ 387
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.081 | 336 | 386 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.96E-14 | 339 | 399 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.05E-10 | 340 | 391 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.1E-13 | 341 | 399 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-10 | 342 | 392 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.7E-7 | 342 | 387 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0001228 | Molecular Function | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MNLLYSSSFK YSDGELRKSQ EFMDLNPYHY HQQQQQIQQN SGLMRYRSAP SSILESLVNG 60 TSGHDGGGIE SGDYRYLRSS SPEMDTMLAR FMSSCNGSGD SSSQNLQEFG ERPAIKQEGG 120 DSEMVYQSLP GHNLVTDNSV SVGNSMDSAF NVMSSMALEN SMQATKMSTA NGSNLARQNS 180 SPAGLFSDLG VDNGGSFRAG NGTNGEASPT NKLRRHVNFS SGQRMLPQIA EIGEECIGGR 240 SPEGDVSEAR YMSRFTSDSW DGASLSGLKR QRDNDGNMFS GLNTLDNQDG NSGNRVTGLT 300 HHLSLPKTLS ETATIEKFLD FQGNSVPCKI RAKRGFATHP RSIAERVRRT RISERMRKLQ 360 ELFPNMDKQT NTADMLDLAV EHIKDLQKQV KTLTDTKAKC TCSSKQKHYS SPSA* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
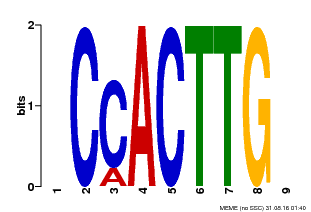
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.009G064700.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024463710.1 | 0.0 | transcription factor bHLH130 isoform X3 | ||||
| TrEMBL | B9HNL6 | 0.0 | B9HNL6_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0009s06910.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 8e-71 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.009G064700.3 |
| Entrez Gene | 7475051 |



