 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.010G040000.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 213aa MW: 24045.2 Da PI: 7.8882 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 33.8 | 6e-11 | 125 | 172 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k Ka +L + ++YI+sLq
Potri.010G040000.2 125 SHSLAERARREKISERMKILQDLVPGC----NKVIGKALVLDEIINYIQSLQ 172
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.79E-18 | 119 | 190 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.39E-8 | 119 | 176 | No hit | No description |
| PROSITE profile | PS50888 | 15.828 | 121 | 171 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.3E-18 | 121 | 190 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.6E-8 | 125 | 172 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.0E-10 | 127 | 177 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048446 | Biological Process | petal morphogenesis | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 213 aa Download sequence Send to blast |
MDPPARMNEG GPYSLEEIWQ FPINGSGRGH FDLLNLEQRA AASVRKRRDV DLDVDDDSSS 60 KPTTNGLLRL KTSGSKDEDH HRHHDSKDEA EPSSGKHVEH KTQPPEPSKQ DYIHVRARRG 120 QATDSHSLAE RARREKISER MKILQDLVPG CNKVIGKALV LDEIINYIQS LQRQVEFLSM 180 KLEAVNTRMN PGIEVFASKD HIINLKVFIK LV* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pth.8562 | 0.0 | leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Isoform 1 is specifically expressed in flowers, mostly in petals, inflorescence and flower buds. Isoform 2 is expressed ubiquitously (leaves, flowers and stems). {ECO:0000269|PubMed:16902407}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the control of petal size, by interfering with postmitotic cell expansion to limit final petal cell size. {ECO:0000269|PubMed:16902407}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00207 | DAP | Transfer from AT1G59640 | Download |
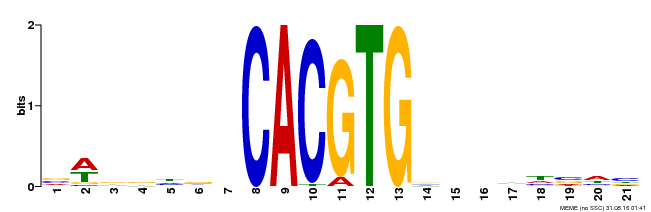
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.010G040000.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Isoform 1 is up-regulated by PI/AP3, SEP2, SEP3 and AP1, but repressed by AG. {ECO:0000269|PubMed:16902407}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024465343.1 | 1e-148 | transcription factor BHLH089 isoform X4 | ||||
| Swissprot | Q0JXE7 | 8e-71 | BPE_ARATH; Transcription factor BPE | ||||
| TrEMBL | A0A3N7H7N7 | 1e-155 | A0A3N7H7N7_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0010s04920.1 | 1e-146 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59640.1 | 4e-71 | BIG PETAL P | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.010G040000.2 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



