 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.015G048000.2 | ||||||||
| Common Name | POPTR_0015s03550g | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 560aa MW: 60871.6 Da PI: 8.8223 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 47.4 | 3.4e-15 | 255 | 304 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr l+P+ ++K +Ka+ L +++e+++ Lq
Potri.015G048000.2 255 RSKHSATEQRRRSKINDRFQMLRALIPHG----DQKRDKASFLLEVIEHVQFLQ 304
889*************************7....7******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 4.2E-24 | 250 | 327 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.23E-18 | 251 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.891 | 253 | 303 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.2E-12 | 255 | 304 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.12E-14 | 255 | 308 | No hit | No description |
| SMART | SM00353 | 2.7E-12 | 259 | 309 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 560 aa Download sequence Send to blast |
MFLTGRKPTH DFLSLYSHSS TVQQDPRPPS QGGFLQTHDF LQPLEQVSKA TAREETNVEI 60 LTIEKPPPPA PPPSVEHTLP GGIGTYSISH VSYFNQRVPK PENTIFSVAQ ASSTDKNDEN 120 SNCSSYSASG FTLWEESTLK KGKTGKENVG ERSNIIREAA AKTDQWTTSE RPSQSSSNNH 180 RNSFSSLSSS QPPGLKCTQS FIEMIKSAKG SNLDDDLDDE ETFLLKKETP SPIHKGELRV 240 KVDGKSNDQK PNTPRSKHSA TEQRRRSKIN DRFQMLRALI PHGDQKRDKA SFLLEVIEHV 300 QFLQEKVQKY EGSYQGWNHE HAKLGPWRNN SRPVESSVDQ SRGVNSGVGP ALLFAANLDE 360 KSITISPSIN PGGARNAVES NMSSASTFNA MDHHPNLGIT NKAMPFPISL QPNLFHPGRI 420 AGAAAQFPPR LAFDAENTAT QPQPCHAISC TSDGAVASDK LKQQNLTVEG GTISISTAYS 480 QGLLNTLTQA LQSSGVDLSQ ATISVQIELG KKGNSRQTAP TSIVKDNNVP PSNQGTIRSR 540 VSSGEESDQA LKKLKTSKG* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Pth.12856 | 0.0 | bud| catkin| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed constitutively in roots. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
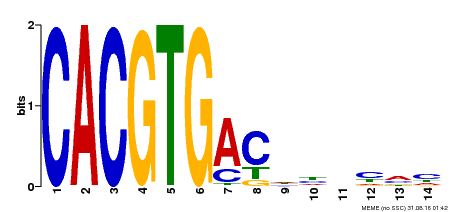
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.015G048000.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EF145375 | 0.0 | EF145375.1 Populus trichocarpa clone WS01123_N13 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024442672.1 | 0.0 | transcription factor BIM1 isoform X2 | ||||
| Swissprot | Q9LEZ3 | 1e-121 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | A0A2K1XI27 | 0.0 | A0A2K1XI27_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0015s03550.1 | 0.0 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.5 | 1e-121 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.015G048000.2 |
| Entrez Gene | 7453864 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



