 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Potri.019G079900.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Salicaceae; Saliceae; Populus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 262aa MW: 28431.7 Da PI: 6.0844 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 34.2 | 4.5e-11 | 197 | 242 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L+el+P+ +k + a++L +A Y+k Lq
Potri.019G079900.3 197 SIAERVRRTRISDRIRKLQELVPNM----DKQTNTADMLDEALAYVKFLQ 242
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.132 | 191 | 241 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.28E-16 | 194 | 255 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.16E-11 | 195 | 246 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-15 | 196 | 255 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.7E-8 | 197 | 242 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.9E-12 | 197 | 247 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0001228 | Molecular Function | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 262 aa Download sequence Send to blast |
MQPTPAGSSN TSAGGGSAGG GGGGGIARFR SAPATWIEAL LGEEEEDPLK QSQNLTELLT 60 SNTPSSRDSV PFNASSAAVE PGLFEPVGGF QRQNSSPTDF LRSSGIGSDQ GYFSSYGNAS 120 NYEYMTPNMD VSPSDKRARE VELQNPSARY PPPPSKGEQT GPLRASSLIE MEMDKLLEDS 180 VPCRVRAKRG CATHPRSIAE RVRRTRISDR IRKLQELVPN MDKQTNTADM LDEALAYVKF 240 LQRQIQELTE QQRKCKCIAK E* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed constitutively in roots, leaves, stems, and flowers. {ECO:0000269|PubMed:12679534}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00181 | DAP | Transfer from AT1G35460 | Download |
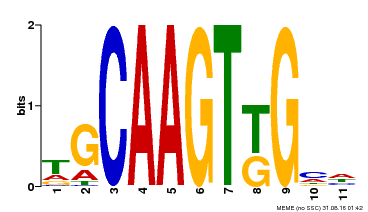
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Potri.019G079900.3 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002325537.3 | 0.0 | transcription factor bHLH81 | ||||
| Swissprot | Q9C8P8 | 1e-85 | BH080_ARATH; Transcription factor bHLH80 | ||||
| TrEMBL | A0A2K1WRC8 | 0.0 | A0A2K1WRC8_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0019s10900.1 | 1e-162 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G35460.1 | 6e-76 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Potri.019G079900.3 |



