 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c11_15370V3.7.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 583aa MW: 65483.8 Da PI: 5.975 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 43.6 | 5.2e-14 | 389 | 435 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h ++Er+RR+++N++f Lr+++P+ K++Ka+ Le A+ YI++Lq
Pp3c11_15370V3.7.p 389 NHVQAERQRREKLNQKFYALRSVVPNV-----SKMDKASLLEDAITYINELQ 435
6999**********************6.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.9E-41 | 51 | 194 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| CDD | cd00083 | 1.93E-15 | 383 | 439 | No hit | No description |
| PROSITE profile | PS50888 | 17.352 | 385 | 434 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.63E-19 | 385 | 454 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.4E-19 | 389 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.9E-11 | 389 | 435 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-17 | 391 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009269 | Biological Process | response to desiccation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009963 | Biological Process | positive regulation of flavonoid biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0043619 | Biological Process | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051090 | Biological Process | regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:2000068 | Biological Process | regulation of defense response to insect | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 583 aa Download sequence Send to blast |
MEMSQLLNAW EVADSAMIEA FMGTGYNCVE GFEVQDDPDG QLHLNESVLL RRLHSLVEES 60 TVDWTYAIFW QLSALREGEM MLGWGDGYFR SAKENEINDA RNMKGGSQEE DQQMRRKVLR 120 ELQALVNGSE DDVSDYVTDT EWFYLVSMSH SYAAGVGTPG RALASDRPVW LIGANKAPDN 180 NCSRVQLAKM AGIQIRESWE VVQNVKMVFD EPMMWAAHEI QAVAHSLPLS SDATSMRPSS 240 PSLMSIATIS ASISNASQIG RCSNPSQDHE AHFVGRKNPP SSRPPMRFYK PRVEELSSDT 300 PETNLVDNKY IVGKSVSFHH LSKIGQRGMP GPPTTANRLP CFAPSEVDCH ESEADVSVKE 360 NVVESSTNLE PKPRKRGRKP ANDREEPLNH VQAERQRREK LNQKFYALRS VVPNVSKMDK 420 ASLLEDAITY INELQEKLQK AEAELKVFQR QVLASTGESK KPNPSRRDST ESSDEERFRL 480 QESGQRSAPL VHTSENKPVI SVFVLGEEAM IRVYCTRHSN FIVHMMSALE KLRLEVIHSN 540 TSSMKDMLLH VVIVKIDSTQ TYTQEQLGKI LERSYVPDHS AT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rru_A | 3e-30 | 13 | 194 | 13 | 197 | Transcription factor MYC3 |
| 4ywc_A | 3e-30 | 13 | 194 | 13 | 197 | Transcription factor MYC3 |
| 4ywc_B | 3e-30 | 13 | 194 | 13 | 197 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
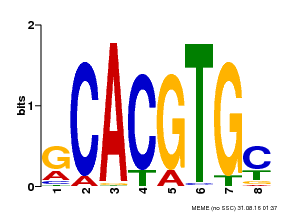
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00084 | PBM | Transfer from AT1G32640 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024388254.1 | 0.0 | transcription factor MYC3-like isoform X1 | ||||
| Refseq | XP_024388255.1 | 0.0 | transcription factor MYC3-like isoform X1 | ||||
| TrEMBL | A9RIS2 | 0.0 | A9RIS2_PHYPA; Predicted protein | ||||
| STRING | PP1S11_350V6.2 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 1e-47 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c11_15370V3.7.p |



