 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c11_21140V3.2.p | ||||||||
| Common Name | Glk2, PHYPADRAFT_178653 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 518aa MW: 56232.6 Da PI: 4.694 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 86.2 | 3.2e-27 | 226 | 280 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k++++WtpeLH+rFv+aveqL G ekA P++ilelm+v++Lt+++++SHLQkYR++
Pp3c11_21140V3.2.p 226 KAKVDWTPELHRRFVHAVEQL-GVEKAYPSRILELMGVQCLTRHNIASHLQKYRSH 280
6799*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.533 | 223 | 282 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-27 | 224 | 283 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 7.17E-18 | 224 | 282 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.8E-25 | 226 | 280 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.5E-7 | 229 | 278 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000006 | anatomy | plant protoplast | ||||
| PO:0025017 | anatomy | plant spore | ||||
| PO:0030003 | anatomy | protonema | ||||
| PO:0030018 | anatomy | gametophore | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 518 aa Download sequence Send to blast |
MAMDMARIDE STAVEVNSLS LVHCVLDGLP DSPCLKSSPT SFEEAVAEGR SVFGDEEDII 60 NNSNDQDNSS SCGAVVTTHE DFAECLNFVT EAECGDVGVR CFEDFDKLPD CGDEGETSKA 120 EEEGCVRIGG GGEQGELLES VSLDCSRNSE NLELRDLGEL WEGSERPDSV PGNEVGEEEA 180 LLLAEAAKAT GDVVSASDSG ECSSVDRKDN QASPKSSKNA APGKKKAKVD WTPELHRRFV 240 HAVEQLGVEK AYPSRILELM GVQCLTRHNI ASHLQKYRSH RRHLAAREAE AASWTHRRTY 300 TQAPWPRSSR RDGLPYLVPI HTPHIQPRPS MAMAMQPQLQ TPHHPISTPL KVWGYPTVDH 360 SNVHMWQQPA VATPSYWQAA DGSYWQHPAT GYDAFSARAC YSHPMQRVPV TTTHAGLPIV 420 APGFPDESCY YGDDMLAGSM YLCNQSYDSE IGRAAGVAAC SKPIETHLSK EVLDAAIGEA 480 LANPWTPPPL GLKPPSMEGV IAELQRQGIN TVPPSTC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-16 | 223 | 278 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.8600 | 0.0 | gametophore| protonema | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
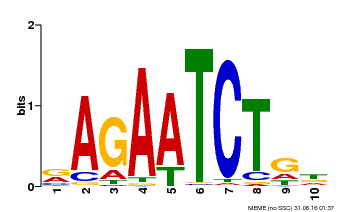
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY741685 | 0.0 | AY741685.1 Physcomitrella patens golden 2-like protein 2 (Glk2) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024388769.1 | 0.0 | transcription activator GLK1-like | ||||
| TrEMBL | Q5IFL6 | 0.0 | Q5IFL6_PHYPA; Golden 2-like protein 2 | ||||
| STRING | PP1S31_317V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 2e-42 | GBF's pro-rich region-interacting factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c11_21140V3.2.p |
| Entrez Gene | 5921460 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



