 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c11_23440V3.1.p | ||||||||
| Common Name | PHYPADRAFT_120051 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 337aa MW: 37452.5 Da PI: 4.6915 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.2 | 2.8e-18 | 89 | 142 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t +q++ Le+ Fe ++++ e++ +LAk+lgL+ rqV vWFqNrRa++k
Pp3c11_23440V3.1.p 89 KKRRLTFDQVRSLERNFEVENKLEPERKMQLAKELGLQPRQVAVWFQNRRARWK 142
45578999*********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 124.6 | 4.5e-40 | 88 | 179 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ +qv++LE++Fe e+kLeperK++la+eLglqprqvavWFqnrRAR+ktkqlE+dye+L + y++lk+e e++ +e++eL+ e+
Pp3c11_23440V3.1.p 88 EKKRRLTFDQVRSLERNFEVENKLEPERKMQLAKELGLQPRQVAVWFQNRRARWKTKQLERDYEVLTSDYNRLKSEFEAVLQEKQELQGEI 178
69*************************************************************************************9988 PP
HD-ZIP_I/II 92 k 92
+
Pp3c11_23440V3.1.p 179 E 179
6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.24E-19 | 79 | 146 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.589 | 84 | 144 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.7E-14 | 87 | 148 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.85E-16 | 89 | 145 | No hit | No description |
| Pfam | PF00046 | 1.2E-15 | 89 | 142 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-19 | 91 | 151 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 6.0E-6 | 115 | 124 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 119 | 142 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 6.0E-6 | 124 | 140 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.5E-16 | 144 | 186 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0030018 | anatomy | gametophore | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MANNISRGGF GHGSSNVMLV RNESSTDTLV AMLASCSPAA LQVQRGGGGL EDALVSSGQK 60 RSFFPTFEAS GEDAGDEDLG DDCTHNVEKK RRLTFDQVRS LERNFEVENK LEPERKMQLA 120 KELGLQPRQV AVWFQNRRAR WKTKQLERDY EVLTSDYNRL KSEFEAVLQE KQELQGEIEC 180 LTGKLQISSQ PGPVDGAPEK PSKRLSKAVP SQLSPPKTSS EACVKSERKP SDPNASSNDD 240 KEEGSRATSS DSISSEILDA DSPRTTEKFN TDLAAEVPLD SNIIHVSDHL CDHMLYPQAC 300 QRISVKLEDG SFRDESCNYI LSQLDEERGL PWWDWP* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 136 | 144 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
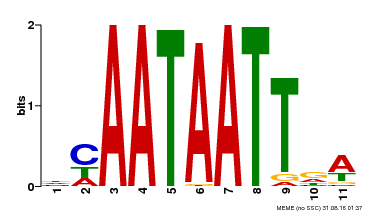
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024388198.1 | 0.0 | homeobox-leucine zipper protein HAT5-like | ||||
| TrEMBL | A0A2K1JVY6 | 0.0 | A0A2K1JVY6_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S31_96V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G01470.1 | 3e-45 | homeobox 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c11_23440V3.1.p |
| Entrez Gene | 5921442 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



