 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c12_17450V3.13.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 452aa MW: 47757.5 Da PI: 7.0801 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 41.8 | 2.5e-13 | 44 | 88 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
Pp3c12_17450V3.13.p 44 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 88
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.64E-16 | 38 | 94 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 39 | 93 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-6 | 42 | 94 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.3E-18 | 42 | 91 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-11 | 43 | 91 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.2E-11 | 44 | 88 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.79E-8 | 46 | 89 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 452 aa Download sequence Send to blast |
MNAASVEMLG VMVAGSTVPG IPSVSVSEEG SKKIRKPYTI TKSRESWTEQ EHDKFLEALQ 60 LFDRDWKKIE AFVGSKTVIQ IRSHAQKYFL KVQKNGTGEH VPPPRPKRKS AQPYPQKAPK 120 CVPAQVQVSD GLNAQAVQAE SSYGTGSHVP PMSSASPSVS AWVQHSVSPN PSISYDAPGI 180 KSEVEGVSLT AVRASSNSIS GSSPGGWPQH VLPASQIAPE SCIRAAPDFT EVYKFIGSVF 240 DPGVSGHLRK LKEMSPIDRE TVLLLMRNLS INLASPDFEE HKLYLSVYVN KEIKGKSTGT 300 QDLTSAPSAA AQSSSQDTGE DTSSVSGQAG TPQSNALLVS PRSGDADHTQ QRASLLLAYS 360 DNTADAGTPR RHSYDGGGAI SASYTSSGVS PSPGDLEPVS LGPPSAALQP QIPVLTMKPE 420 RVYSAPSLPS LTSLPSLPAG LVDGSVDTWW T* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.14160 | 1e-163 | gametophore| protonema | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
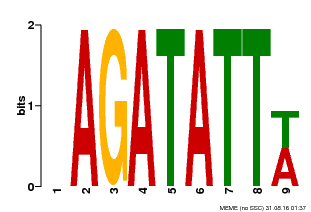
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024389902.1 | 0.0 | protein REVEILLE 3-like isoform X1 | ||||
| Refseq | XP_024389903.1 | 0.0 | protein REVEILLE 3-like isoform X1 | ||||
| Refseq | XP_024389904.1 | 0.0 | protein REVEILLE 3-like isoform X1 | ||||
| Refseq | XP_024389905.1 | 0.0 | protein REVEILLE 3-like isoform X1 | ||||
| Refseq | XP_024389906.1 | 0.0 | protein REVEILLE 3-like isoform X1 | ||||
| Swissprot | Q6R0H0 | 3e-87 | RVE3_ARATH; Protein REVEILLE 3 | ||||
| TrEMBL | A0A2K1KLY0 | 0.0 | A0A2K1KLY0_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S46_272V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP928 | 15 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01280.2 | 2e-82 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c12_17450V3.13.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



