 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c13_1060V3.6.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 190aa MW: 20672.7 Da PI: 10.0619 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 39.9 | 1e-12 | 5 | 55 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
+++WT+eE+ l +v+++G+g W++I + + R+ ++k++w++
Pp3c13_1060V3.6.p 5 KQKWTAEEEAALRAGVEKYGPGKWRAIQKDSKfgpclTSRSNVDLKDKWRN 55
79************************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.36 | 1 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.22E-16 | 2 | 58 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-9 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.4E-10 | 5 | 55 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-15 | 5 | 56 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 4.45E-22 | 6 | 56 | No hit | No description |
| SMART | SM00526 | 1.2E-12 | 115 | 180 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 20.416 | 117 | 189 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 3.48E-13 | 119 | 180 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 3.2E-13 | 119 | 178 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 4.7E-10 | 120 | 178 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0006357 | Biological Process | regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003691 | Molecular Function | double-stranded telomeric DNA binding | ||||
| GO:0033613 | Molecular Function | activating transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043047 | Molecular Function | single-stranded telomeric DNA binding | ||||
| GO:0070491 | Molecular Function | repressing transcription factor binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 190 aa Download sequence Send to blast |
MGAPKQKWTA EEEAALRAGV EKYGPGKWRA IQKDSKFGPC LTSRSNVDLK DKWRNMSVSA 60 NGLGSARKPL AITGGPGMMT LMEDAVSVLP LAVLPPIDDA QALKRESADT SGDRKSLGSR 120 YDDMVFEAVI GLKEPYGSSN ASITSYIEER HAVPSNFRRL LTTKLKELAL AGKLVKVRYS 180 WKHEIANAH* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.11660 | 0.0 | gametophore| protonema| sporophyte | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
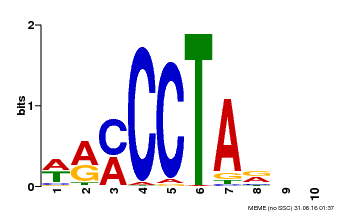
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024393158.1 | 1e-128 | single myb histone 1-like | ||||
| TrEMBL | A9T749 | 1e-123 | A9T749_PHYPA; Single myb histone protein | ||||
| STRING | PP1S176_113V6.2 | 1e-119 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 2e-36 | telomere repeat binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c13_1060V3.6.p |



