 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c13_15030V3.12.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 865aa MW: 93696.9 Da PI: 5.5546 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 40.7 | 5e-13 | 357 | 416 | 2 | 61 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklks 61
+ kr r+++NRe+A+ sRqRKk++++eLe k +++a +L ++++l+ e +l+
Pp3c13_15030V3.12.p 357 DDEKRRARLMRNRESAQLSRQRKKMYVDELEGKLRTMTATVAELNATISHLTAENVNLRR 416
56799***********************************************99888876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.5E-15 | 356 | 420 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.461 | 358 | 421 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.0E-10 | 359 | 417 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 9.2E-16 | 360 | 418 | No hit | No description |
| SuperFamily | SSF57959 | 1.0E-11 | 360 | 418 | No hit | No description |
| CDD | cd14704 | 5.78E-21 | 361 | 411 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006990 | Biological Process | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response | ||||
| GO:0009408 | Biological Process | response to heat | ||||
| GO:0005635 | Cellular Component | nuclear envelope | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| GO:0005783 | Cellular Component | endoplasmic reticulum | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0048471 | Cellular Component | perinuclear region of cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 865 aa Download sequence Send to blast |
MSFQNMERKI EIPDIRGDDG ETLEKEDPEI ATLSSSEASV NWENISEDWV SLFLDPATFD 60 EHATPLPSDL ESYSRFDSTL RESNDRFSPQ ELLADSLLLH SIDDQLSSDL VLQSSVNVVE 120 DIFGKDGSST KGLKRADLKH SEGEPRGQNS ATVSNFSPAP QSNATQTACA STSMEVYASS 180 SPASSLTQVS GDSSPGMEVD LWKNKEANSD LLITLEQRVN GEVSSRSESP TSPSSAMGVE 240 NGCPNVKKLQ SEGGRTGSRD VSSTGSAVRT GIPSTALQSD AFDVRQLRSI DNSFKVGFLG 300 RSGDTEDSVE DREESNKLFC REYFQSESFK LQNLEVPEDQ QSDRSDGERK ADAGVEDDEK 360 RRARLMRNRE SAQLSRQRKK MYVDELEGKL RTMTATVAEL NATISHLTAE NVNLRRQLGY 420 YYPAPGVCPP RPGMPMQVLP MGPYPGMVAG RPIYSGGQMP PVPIPRIKAK IPARSTKRAK 480 SEDTKEGGDR KRTKLKAAGA ASMAVMGLLC VAMLFSSFDQ GLNGSGGVED YTRIGNVRIG 540 GRVLTSWDEA VNPLNNTGLS SSDTEPGWVP LQEGEYGPER EGVANIHMRL AAEKVQDEAV 600 KSQISKAGLE SSARQSVDGN EDLGISKEKG SQLPPDYFSS GKVPLQMRRE IVSTNTSQSY 660 AATVFVPGAN GLVKVDGNLI IQAVMAGDKA AKKQSKNKQI VGGKKKGPVS KNLPVKVLDN 720 LAPRETKVVQ NATTGPFVIA RPLPERAKSV FGTNNPVSPV LHAGALQQWM LGGLHDSETN 780 FIASELTKIM SQPRTQLPID NSEAVAATQR TYHTFTLYDA APRGPLHSWL FAGLSTGAVH 840 VVTSACNMVY VIHTSRNFLL QINI* |
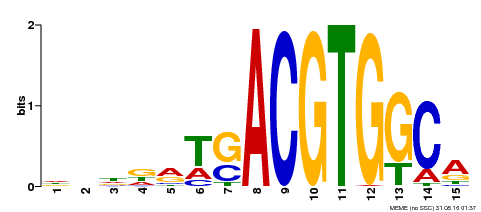
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00345 | DAP | Transfer from AT3G10800 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024392783.1 | 0.0 | uncharacterized protein LOC112290591 isoform X5 | ||||
| TrEMBL | A0A2K1JM11 | 0.0 | A0A2K1JM11_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S158_117V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10800.1 | 1e-32 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c13_15030V3.12.p |



