 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c16_15450V3.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 855aa MW: 93170.2 Da PI: 6.2622 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.2 | 5.8e-18 | 27 | 85 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le++++++++ps +r +L k++ +++ +q+kvWFqNrR +ek+
Pp3c16_15450V3.2.p 27 KYVRYTNEQVEALERVYHECPKPSSIRRHQLIKECpilaNIEPKQIKVWFQNRRCREKQ 85
5678*****************************************************97 PP
| |||||||
| 2 | START | 157.6 | 9.9e-50 | 185 | 392 | 2 | 204 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..ga 90
+aee+++++++ka+ ++ W +++ +++g++ + +++ s+++ g a+ra+g+ + v+e+l+d++ W+++++++e+l ++ +g g+
Pp3c16_15450V3.2.p 185 MAEETLTDFLAKATGTAVDWIQLPGMKPGPDAIGIIAISHGCVGIAARACGLAALDFS-KVAEILKDRPGWSQDCRRMEVLGTLPTGngGT 274
57999*****************************************************.8888888888********************** PP
EEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX CS
START 91 lqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlp 177
++l +++++a+++l+p Rdf ++Ry+ l++g +vi+++S++ + + p+ +s++Rae++pSg+li+p+++g++ +++v+hv+++ +++
Pp3c16_15450V3.2.p 275 IELLYTQMYAPTTLAPaRDFCTLRYTTLLEDGNLVICERSLTGKHNGPTmppVQSFIRAEMFPSGYLIRPCEGGGCIIHIVDHVEYEPWSV 365
******************************************999988888899************************************* PP
HHHHHHHHHHHHHHHHHHHHHHTXXXX CS
START 178 hwllrslvksglaegaktwvatlqrqc 204
+++lr+l++s ++ + k + a+l++++
Pp3c16_15450V3.2.p 366 PEVLRPLYESPAVLAHKSTIAALRYLR 392
**********************99875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.499 | 22 | 86 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.87E-17 | 22 | 89 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 8.9E-15 | 24 | 90 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.56E-16 | 27 | 87 | No hit | No description |
| Pfam | PF00046 | 1.7E-15 | 28 | 85 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-18 | 29 | 85 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 0.001 | 79 | 115 | No hit | No description |
| PROSITE profile | PS50848 | 22.643 | 175 | 375 | IPR002913 | START domain |
| CDD | cd08875 | 1.81E-79 | 179 | 395 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 2.4E-20 | 184 | 375 | IPR023393 | START-like domain |
| SMART | SM00234 | 3.1E-41 | 184 | 394 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.33E-34 | 185 | 395 | No hit | No description |
| Pfam | PF01852 | 4.4E-47 | 186 | 392 | IPR002913 | START domain |
| Pfam | PF08670 | 1.4E-45 | 713 | 854 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 855 aa Download sequence Send to blast |
MAGERCFEAS SNLVDPDKYK GMDSSGKYVR YTNEQVEALE RVYHECPKPS SIRRHQLIKE 60 CPILANIEPK QIKVWFQNRR CREKQRKEAT RLVSVNAKLT ALNKLLMEEN ERLAKHASQL 120 TLDNHALRQQ LPNLPVPDGK CRLSGQAGVA STDTSCDSAV TGGLPQHLTS QHSSPDASPA 180 GLLSMAEETL TDFLAKATGT AVDWIQLPGM KPGPDAIGII AISHGCVGIA ARACGLAALD 240 FSKVAEILKD RPGWSQDCRR MEVLGTLPTG NGGTIELLYT QMYAPTTLAP ARDFCTLRYT 300 TLLEDGNLVI CERSLTGKHN GPTMPPVQSF IRAEMFPSGY LIRPCEGGGC IIHIVDHVEY 360 EPWSVPEVLR PLYESPAVLA HKSTIAALRY LRRIAAEESG EIIIRNGQHP AVIRTLSQRL 420 TKGFNDAVNG FGDDGWVPME SDGMDDVSVM LNATPKSMEG QIATDKLLFS LGGGILCAKA 480 SMLLQNVPPA LLIRFLREHR SEWADHEIDA NAATAFRGAS NGHVSRGRMS HVQLPLPLAQ 540 FGEQGEFLEV VKLEGHSAVQ HSVLSRDSFL LQLCSGIEEG AVGAGAQLVF APIDAAVSED 600 IPLLPSGFRV IPVDSSVVDG IGLNRTLDLA STLEDHEAGL NGESKSNGSS SQVRSVLTIA 660 FQFAYEVHTR ETCAVMARQY VRTVVASVQR VAMALAPSRG QPRPALGNSD AISLARHILS 720 SYRVQLGMDL VRPEVGGTEA LFKVFWHHSD AIVCCAWKGT PEFVFANRAG LEMFETTSSS 780 LQDLAWDKTL DEDSLKLSYA TFTQVLQQGY CCLPSGVRIS STGRTVTYER ALAWKVLDDD 840 ETTQCIAFLF MNWS* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.16218 | 0.0 | gametophore | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, roots, stems, leaf sheaths and blades and panicles. {ECO:0000269|PubMed:17999151}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
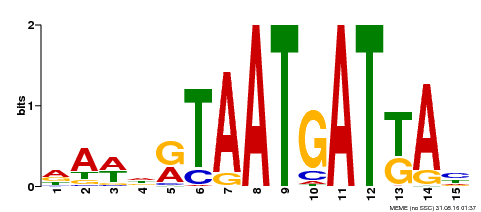
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ657202 | 0.0 | DQ657202.1 Physcomitrella patens class III HD-Zip protein HB12 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024399219.1 | 0.0 | homeobox-leucine zipper protein HOX32-like isoform X2 | ||||
| Swissprot | Q6AST1 | 0.0 | HOX32_ORYSJ; Homeobox-leucine zipper protein HOX32 | ||||
| TrEMBL | Q0Q429 | 0.0 | Q0Q429_PHYPA; Class III HD-Zip protein HB12 | ||||
| STRING | PP1S15_11V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.2 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c16_15450V3.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



