 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c19_15710V3.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 477aa MW: 51014.1 Da PI: 7.6229 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.2 | 1.1e-10 | 295 | 353 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i eLe+kv+ L++e ++L +l +l+k l +e+
Pp3c19_15710V3.1.p 295 KRAKRILANRQSAARSKERKMRYISELERKVQGLQTEATTLSTQLAMLQKDTTGLATEN 353
9************************************************9887776665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.0E-16 | 291 | 355 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.921 | 293 | 356 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 9.0E-10 | 295 | 353 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.99E-11 | 295 | 347 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.4E-11 | 295 | 350 | No hit | No description |
| CDD | cd14703 | 1.72E-22 | 296 | 345 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 477 aa Download sequence Send to blast |
MEGLGKGYDA FVERLQSEFG ATSVGKGSGS SSTSNIAAHP PHPSQSQLHQ SRYVGYPSTS 60 HPFTVKRETS PSVSETSMRS RESYSLEVNM QDAVTSASPA SISGSGQPPR MPSPGHNFST 120 DVNQMPDTPP RRRGHRRAQS EIAFRLPDDA SFESELGVHG SEMPTLSDDG AEDLFSMYID 180 MEQINNMSGT SGQAGAKAGG EGSNAPAPSA HHARSLSADG SLGNLAGNRT GVGVGGGGGN 240 NSAPSEARRP RHGHSSSMDG STSFRHDLLS GDFEGDTKKV MASAKLSEIA LIDPKRAKRI 300 LANRQSAARS KERKMRYISE LERKVQGLQT EATTLSTQLA MLQKDTTGLA TENNELKLRL 360 QAMEQQAHLR DALNEALREE VQRLKVATGQ ISNGSVQNLS MGGQHLFQMQ NQAFNSQQLQ 420 QAQSGLNNPA QQQQQASQEQ MHSEYMQRGA YNLSSGFIKN EGPSIAVKHA SSASFG* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.9872 | 0.0 | gametophore| protonema | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed constitutively at a low level in young seedlings and in roots, stems and leaves of mature plants. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
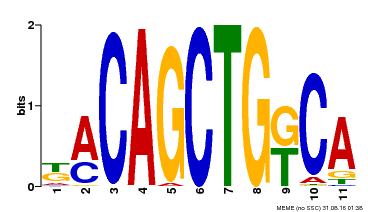
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024356505.1 | 0.0 | transcription factor RF2a-like | ||||
| Refseq | XP_024356506.1 | 0.0 | transcription factor RF2a-like | ||||
| Swissprot | Q04088 | 3e-85 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A2K1IYK4 | 0.0 | A0A2K1IYK4_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S20_118V6.1 | 0.0 | (Physcomitrella patens) | ||||
| STRING | PP1S20_119V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP126 | 17 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 9e-90 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c19_15710V3.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



