 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c1_6880V3.3.p | ||||||||
| Common Name | e2f, PHYPADRAFT_122002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 401aa MW: 44124.4 Da PI: 4.8344 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 86.3 | 2.3e-27 | 74 | 139 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r+++sL+llt+kf++l++++++g+++ln++a++L +v ++RRiYDi+NVLe+++liekk kn+irwkg
Pp3c1_6880V3.3.p 74 RYDSSLGLLTKKFIDLIKQADDGVLDLNKAADTL---HV--QKRRIYDITNVLEGIGLIEKKLKNRIRWKG 139
6899******************************...99..****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 4.4E-29 | 70 | 140 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 8.71E-18 | 74 | 137 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.8E-34 | 74 | 139 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 9.2E-25 | 76 | 139 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| CDD | cd14660 | 2.62E-45 | 150 | 255 | No hit | No description |
| SuperFamily | SSF144074 | 1.49E-30 | 154 | 255 | No hit | No description |
| Pfam | PF16421 | 1.5E-31 | 157 | 254 | IPR032198 | E2F transcription factor, CC-MB domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008284 | Biological Process | positive regulation of cell proliferation | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010090 | Biological Process | trichome morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0051446 | Biological Process | positive regulation of meiotic cell cycle | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000006 | anatomy | plant protoplast | ||||
| PO:0025017 | anatomy | plant spore | ||||
| PO:0030003 | anatomy | protonema | ||||
| PO:0030018 | anatomy | gametophore | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 401 aa Download sequence Send to blast |
MKRRNECEEH HVEMNGWNGY TNSDLSPAPT PTGPRGRGSR AKTVKQTKNG PQTPGPSGIG 60 SPTSSAPTPT STCRYDSSLG LLTKKFIDLI KQADDGVLDL NKAADTLHVQ KRRIYDITNV 120 LEGIGLIEKK LKNRIRWKGL GMVRNAEAKD DAAGLLAEVE DLRIKEKKLD ESISEMRERL 180 RSLSEDEHNK QWLYVTEDDI KNLHCFQNET LIAIKAPLGT TLEVPDPDEA VEYPHRRFQI 240 LLRSTLGPID VYLVSRFEGR TEVPMETLPD SQEAGPSSSV DGMSHQENMV MVPEVGYGLS 300 DLVPPVSHES ASQAPEPTGS HPDFAGGIMR IAPAEVNTDT DYWLLSDAGV GISDMWRSDP 360 SNAMWDEVVR LNPEFGSENI GSPRPHTPPS NNALEVDPVS * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 5e-26 | 69 | 141 | 2 | 75 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.3026 | 0.0 | protonema | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in a cell cycle-dependent manner. Most abundant at the G1/S transition. Lower but constant level in the following phases. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:16514015}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in proliferating cells and several differentiated tissues. Detected in inflorescence and shoot apical meristems, cotyledonary vascular tissues, leaf primordia, young leaves, base of trichomes, central cylinder and elongation zone of roots, lateral root primordia, flowers, pistils of immature flowers and pollen grains. {ECO:0000269|PubMed:16514015}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3' found in the promoter region of a number of genes whose products are involved in cell cycle regulation or in DNA replication. The binding of retinoblastoma-related proteins represses transactivation. Involved in the control of cell-cycle progression from G1 to S phase and from G2 to M phase. Stimulates cell proliferation and delays differentiation. Represses cell enlargement and endoreduplication in auxin-free conditions. {ECO:0000269|PubMed:11669580, ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11862494, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12913157, ECO:0000269|PubMed:16055635, ECO:0000269|PubMed:16514015}. | |||||
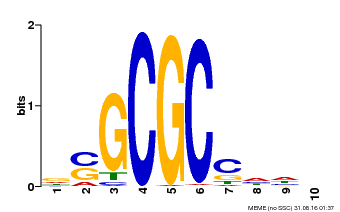
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00661 | PBM | 25215497 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by light and by auxin. May be up-regulated by E2FA. {ECO:0000269|PubMed:16055635, ECO:0000269|PubMed:16514015, ECO:0000269|PubMed:18424613}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ428951 | 0.0 | AJ428951.1 Physcomitrella patens mRNA for putative E2F transcription factor (e2f gene). | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024387480.1 | 0.0 | transcription factor E2FB-like isoform X1 | ||||
| Refseq | XP_024387489.1 | 0.0 | transcription factor E2FB-like isoform X2 | ||||
| Refseq | XP_024387498.1 | 0.0 | transcription factor E2FB-like isoform X3 | ||||
| Refseq | XP_024387507.1 | 0.0 | transcription factor E2FB-like isoform X4 | ||||
| Refseq | XP_024387515.1 | 0.0 | transcription factor E2FB-like isoform X4 | ||||
| Swissprot | Q9FV71 | 1e-103 | E2FB_ARATH; Transcription factor E2FB | ||||
| TrEMBL | Q8GU41 | 0.0 | Q8GU41_PHYPA; Predicted protein | ||||
| STRING | PP1S38_356V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22220.2 | 1e-101 | E2F transcription factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c1_6880V3.3.p |
| Entrez Gene | 5922938 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



