 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c2_23660V3.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 952aa MW: 104023 Da PI: 7.127 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.4 | 8.3e-14 | 259 | 307 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+++GV ++grW A+I++ ++r++lg+f t+eeAa+a++ a+ k++g
Pp3c2_23660V3.2.p 259 SQFRGVV-PQSNGRWGAQIYEK-----HQRIWLGTFNTEEEAARAYDTAAIKFRG 307
6899996.6779*********3.....5************************998 PP
| |||||||
| 2 | B3 | 98.6 | 3.9e-31 | 424 | 524 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldg 86
f+k+ tpsdv+k++rlv+pk++ae++ +++ ++tl++ed sg+ W+++++y+++s++yvltkGW++Fvk+++L++gD+v F++
Pp3c2_23660V3.2.p 424 FDKAVTPSDVGKLNRLVIPKQHAERCfpldlsA-NSP-GQTLSFEDVSGKHWRFRYSYWNSSQSYVLTKGWSRFVKEKKLDAGDIVSFERG- 512
89***************************9853.433.459************************************************43. PP
SSEE..EEEEE-S CS
B3 87 rsefelvvkvfrk 99
++ el++ ++rk
Pp3c2_23660V3.2.p 513 -RNHELYIDFRRK 524
.667779999987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.19E-25 | 259 | 317 | No hit | No description |
| SuperFamily | SSF54171 | 1.18E-17 | 259 | 315 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.5E-21 | 260 | 315 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.521 | 260 | 315 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.1E-30 | 260 | 321 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.6E-5 | 261 | 272 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 7.9E-8 | 261 | 307 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.6E-5 | 281 | 297 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 7.7E-42 | 415 | 525 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.09E-30 | 420 | 524 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.94E-27 | 422 | 523 | No hit | No description |
| PROSITE profile | PS50863 | 15.679 | 424 | 525 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.4E-28 | 424 | 525 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 6.8E-28 | 424 | 524 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 952 aa Download sequence Send to blast |
MCLNAKDEKA VGMWSKPASR SSSEPHHGPD VTLRLNNFDT LVASSSDSDV SSFKIVECPS 60 QGREEASTSV VIRETNPASP IPKFYTIEEK TAMLGVAEAA AANTLQVHGI PRFQRHIDHD 120 RSRLHDSSML RTSGKDSENC FRADAEEHRA YVASNLESFG ERSHPLDLLG SRLEAEFSRG 180 ANLKQWPLGS RNVGQRCGSG ELERTQSGPS AFLGSGSLDT EQRGSLERTH SGPSFSGSGD 240 SEYGDDRGRE QSNSKLPSSQ FRGVVPQSNG RWGAQIYEKH QRIWLGTFNT EEEAARAYDT 300 AAIKFRGRDA MTNFRPVTDS EYESEFLRSF SKEQIVEMLR RHTYDEELDQ CKKVFNMDAA 360 ANPVSARRRV LEMCRSSNPG TRLSVGSFSL LQNTFSADTS TTLAPPNLPR DEPRESSPTR 420 EHLFDKAVTP SDVGKLNRLV IPKQHAERCF PLDLSANSPG QTLSFEDVSG KHWRFRYSYW 480 NSSQSYVLTK GWSRFVKEKK LDAGDIVSFE RGRNHELYID FRRKQITAGG TSTSDRSSFR 540 SAWSGPCNNA YAPSNGPVSP LNSRMWQPFS FSVPHVTVTS SGIQPSSMYG SIYNQMMQSA 600 RGTSAASGDL GSVQDSAIPS MAGSCLNLDL QHSPLKCHAA TEDFILHIDR PLRQSADMDG 660 GSNIHDRSLV ASSSSTSSQS EEAPSTSSGT RLFGVDLERA APLACKVSPL QSIPSFTSSA 720 LQSKLSEGHE SSQGNLKTIH DGGNIPPSAS LSALTSGEFL SFRLGNAFKS LQCKHPVEQS 780 SPENGGAPVR LSLQASTSSS SGHRGSSPLK STAMTEGEDP KARSSTESVA SALDSNSSRS 840 SPMLLEKRKR ENSNRDYMQG CSDQADGAAN VPLTLAHSEF RSSLEQARHA WHKHEDDASA 900 EVQTSVKRHC SPIQASLLRQ GSGTTEQSDF LPKPDLVQEK QKPQYDSSNL R* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5os9_A | 2e-51 | 420 | 524 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| 5os9_B | 2e-51 | 420 | 524 | 4 | 113 | B3 domain-containing transcription factor NGA1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
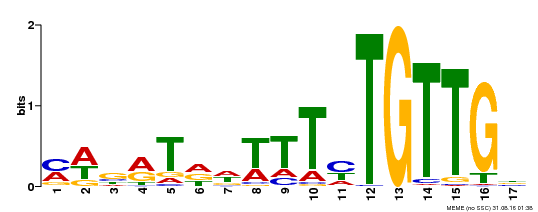
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024369136.1 | 0.0 | uncharacterized protein LOC112279179 isoform X1 | ||||
| Refseq | XP_024369137.1 | 0.0 | uncharacterized protein LOC112279179 isoform X1 | ||||
| TrEMBL | A0A2K1L2S1 | 0.0 | A0A2K1L2S1_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S135_69V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP365 | 16 | 108 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 4e-90 | related to ABI3/VP1 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c2_23660V3.2.p |



